+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
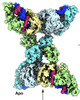
| Title | cryoEM structure of SPARTA complex ligand-free | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SPARTA / TIR / prokaryotic argonaute / ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) | |||||||||
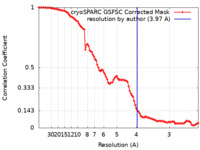
| Method | single particle reconstruction / cryo EM / Resolution: 3.97 Å | |||||||||
 Authors Authors | Babatunde E / Dong CN / Xu HL / Henning S | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Activation mechanism of a short argonaute-TIR prokaryotic immune system. Authors: Dongchun Ni / Xuhang Lu / Henning Stahlberg / Babatunde Ekundayo /   Abstract: Short prokaryotic argonaute (pAgo) and toll/interleukin-1 receptor/resistance protein (TIR)-analog of PAZ (APAZ) form a heterodimeric SPARTA complex that provides immunity to its prokaryotic host ...Short prokaryotic argonaute (pAgo) and toll/interleukin-1 receptor/resistance protein (TIR)-analog of PAZ (APAZ) form a heterodimeric SPARTA complex that provides immunity to its prokaryotic host through an abortive infection mechanism. Monomeric SPARTA senses foreign RNA/DNA duplexes to assemble an active tetramer resulting in cell death by nicotinamide adenine dinucleotide (oxidized form) (NAD) depletion via an unknown mechanism. We report nine structures of SPARTA in different functional states at a resolution range of 4.2 to 2.9 angstroms, revealing its activation mechanism. Inactive SPARTA monomers bind to RNA/DNA duplexes to form symmetric dimers mediated by the association of Ago subunits. The initiation of tetramer assembly induces flexibility of the TIR domains enabling a symmetry-breaking rotational movement of a TIR domain in the dimer units which facilitates the TIR oligomerization, resulting in the formation of the substrate binding pocket and the activation of the SPARTA complex's NADase activity. Our findings provide detailed structural and mechanistic insights into activating a short argonaute defense system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17299.map.gz emd_17299.map.gz | 51.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17299-v30.xml emd-17299-v30.xml emd-17299.xml emd-17299.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17299_fsc.xml emd_17299_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17299.png emd_17299.png | 183 KB | ||
| Masks |  emd_17299_msk_1.map emd_17299_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_17299_additional_1.map.gz emd_17299_additional_1.map.gz emd_17299_half_map_1.map.gz emd_17299_half_map_1.map.gz emd_17299_half_map_2.map.gz emd_17299_half_map_2.map.gz | 53.2 MB 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17299 http://ftp.pdbj.org/pub/emdb/structures/EMD-17299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17299 | HTTPS FTP |
-Validation report
| Summary document |  emd_17299_validation.pdf.gz emd_17299_validation.pdf.gz | 1.4 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17299_full_validation.pdf.gz emd_17299_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  emd_17299_validation.xml.gz emd_17299_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_17299_validation.cif.gz emd_17299_validation.cif.gz | 23.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17299 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17299 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17299 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17299 | HTTPS FTP |
-Related structure data
| Related structure data |  8oz6MC  8ozcC  8ozdC  8ozeC  8ozfC  8ozgC  8oziC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17299.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17299.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17299_msk_1.map emd_17299_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
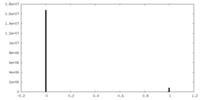
| Density Histograms |
-Additional map: #1
| File | emd_17299_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
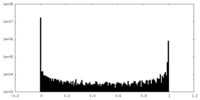
| Density Histograms |
-Half map: #1
| File | emd_17299_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
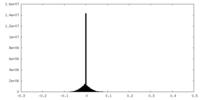
| Density Histograms |
-Half map: #2
| File | emd_17299_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
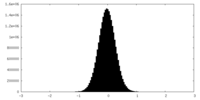
| Density Histograms |
- Sample components
Sample components
-Entire : heter complex of short argonaute-TIR antiviral defence system
| Entire | Name: heter complex of short argonaute-TIR antiviral defence system |
|---|---|
| Components |
|
-Supramolecule #1: heter complex of short argonaute-TIR antiviral defence system
| Supramolecule | Name: heter complex of short argonaute-TIR antiviral defence system type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: TIR domain-containing protein
| Macromolecule | Name: TIR domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 53.270594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRNKIFISHA TPDDNDFTRW LALKLIGLGY EVWCDILFLD KGVDFWSNIE KVIREDTCKF LLVSSSYSNQ REGVLKELAV AAKVKKQLK DDKFIIPLAI DEQLSYDDIN IDIVRLNAID FKMSWARGLK DILEAFEKQK VPKEVADASK SNLLYQQIFL H DKSVIEKE ...String: MRNKIFISHA TPDDNDFTRW LALKLIGLGY EVWCDILFLD KGVDFWSNIE KVIREDTCKF LLVSSSYSNQ REGVLKELAV AAKVKKQLK DDKFIIPLAI DEQLSYDDIN IDIVRLNAID FKMSWARGLK DILEAFEKQK VPKEVADASK SNLLYQQIFL H DKSVIEKE EIYDSNWLSI LSFPEELRFH EYNWMLPKRF DVRELTFPAV RYKNYLCTFA WAYDFTYHLP KTETYHKSKT IR IPTEEIL SGSYDSNFIR NAECKRLIVQ LLNKAFELRM KDKEVQEYEM SNKTAYWLEK GKLEKDKFEK TMLVGKQKDK NWH FAISGA SKLYPFPVLM ISSHIFFTAD GKKLIDSSSV QHSSRRRQGK NWWNNTWRTK LLAFIKYLSD DDTSFYLEMG SEEK VFVSN EPVKFKGNVS YNIPEKNTLE EEAELSGFNQ GEDIEELEEL IENLEAE UniProtKB: TIR domain-containing protein |
-Macromolecule #2: Piwi domain-containing protein
| Macromolecule | Name: Piwi domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 58.09141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKELIYIEEP KILFAHGQKC TDARDGLALF GPLNNLYGIK SGVIGTKQGL KIFRDYLDHI QKPIYNSNSI TRPMFPGFEA VFDCKWEST GITFKEVTNE DIGKFLYNSS THKRTYDLVS LFIDKIISAN KNEDENVDVW FVIVPDEIYK YCRPNSVLPK E MVQTKALM ...String: MKELIYIEEP KILFAHGQKC TDARDGLALF GPLNNLYGIK SGVIGTKQGL KIFRDYLDHI QKPIYNSNSI TRPMFPGFEA VFDCKWEST GITFKEVTNE DIGKFLYNSS THKRTYDLVS LFIDKIISAN KNEDENVDVW FVIVPDEIYK YCRPNSVLPK E MVQTKALM SKSKAKSFRY EPSLFPDINI ELKEQEKEAE TYNYDAQFHD QFKARLLKHT IPTQIFREST LAWRDFKNAF GL PIRDFSK IEGHLAWTIS TAAFYKAGGK PWKLSDVRNG VCYLGLVYKK VEKSKNPRNA CCAAQMFLDN GDGTVFKGEV GPW YNPKNG QYHLEPKEAK ALLSQSLQSY KEQIGEYPKE VFIHAKTRFN HQEWDAFLEV TPKETNLVGV TISKTKPLKL YKTE GDYTI LRGNAYVVNE RSAFLWTVGY VPKIQTALSM EVPNPLFIEI NKGEADIKQV LKDILSLTKL NYNACIFADG EPVTL RFAD KIGEILTAST DIKTPPLAFK YYI UniProtKB: Piwi domain-containing protein |
-Macromolecule #3: RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') type: rna / ID: 3 / Number of copies: 4 |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 5.466027 KDa |
| Sequence | String: UUUUUUUUUU UUUUUUUU |
-Macromolecule #4: DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') type: dna / ID: 4 / Number of copies: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 4.966352 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8oz6: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)