[English] 日本語
 Yorodumi
Yorodumi- EMDB-17091: Small subunit of yeast mitochondrial ribosome in complex with MET... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Small subunit of yeast mitochondrial ribosome in complex with METTL17/Rsm22 (local-masked refined on the head) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondria / assembly / biogenesis / iron-sulfur cluster / 4Fe-4S / RIBOSOME | |||||||||
| Biological species |  | |||||||||
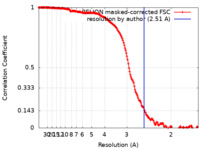
| Method | single particle reconstruction / cryo EM / Resolution: 2.51 Å | |||||||||
 Authors Authors | Itoh Y / Chicherin I / Kamenski P / Amunts A | |||||||||
| Funding support | European Union,  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: METTL17 is an Fe-S cluster checkpoint for mitochondrial translation. Authors: Tslil Ast / Yuzuru Itoh / Shayan Sadre / Jason G McCoy / Gil Namkoong / Jordan C Wengrod / Ivan Chicherin / Pallavi R Joshi / Piotr Kamenski / Daniel L M Suess / Alexey Amunts / Vamsi K Mootha /    Abstract: Friedreich's ataxia (FA) is a debilitating, multisystemic disease caused by the depletion of frataxin (FXN), a mitochondrial iron-sulfur (Fe-S) cluster biogenesis factor. To understand the cellular ...Friedreich's ataxia (FA) is a debilitating, multisystemic disease caused by the depletion of frataxin (FXN), a mitochondrial iron-sulfur (Fe-S) cluster biogenesis factor. To understand the cellular pathogenesis of FA, we performed quantitative proteomics in FXN-deficient human cells. Nearly every annotated Fe-S cluster-containing protein was depleted, indicating that as a rule, cluster binding confers stability to Fe-S proteins. We also observed depletion of a small mitoribosomal assembly factor METTL17 and evidence of impaired mitochondrial translation. Using comparative sequence analysis, mutagenesis, biochemistry, and cryoelectron microscopy, we show that METTL17 binds to the mitoribosomal small subunit during late assembly and harbors a previously unrecognized [FeS] cluster required for its stability. METTL17 overexpression rescued the mitochondrial translation and bioenergetic defects, but not the cellular growth, of FXN-depleted cells. These findings suggest that METTL17 acts as an Fe-S cluster checkpoint, promoting translation of Fe-S cluster-rich oxidative phosphorylation (OXPHOS) proteins only when Fe-S cofactors are replete. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17091.map.gz emd_17091.map.gz | 337 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17091-v30.xml emd-17091-v30.xml emd-17091.xml emd-17091.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17091_fsc.xml emd_17091_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_17091.png emd_17091.png | 73.9 KB | ||
| Masks |  emd_17091_msk_1.map emd_17091_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17091.cif.gz emd-17091.cif.gz | 4.7 KB | ||
| Others |  emd_17091_half_map_1.map.gz emd_17091_half_map_1.map.gz emd_17091_half_map_2.map.gz emd_17091_half_map_2.map.gz | 339.1 MB 339 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17091 http://ftp.pdbj.org/pub/emdb/structures/EMD-17091 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17091 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17091 | HTTPS FTP |
-Validation report
| Summary document |  emd_17091_validation.pdf.gz emd_17091_validation.pdf.gz | 1005.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17091_full_validation.pdf.gz emd_17091_full_validation.pdf.gz | 1005.1 KB | Display | |
| Data in XML |  emd_17091_validation.xml.gz emd_17091_validation.xml.gz | 24.1 KB | Display | |
| Data in CIF |  emd_17091_validation.cif.gz emd_17091_validation.cif.gz | 31.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17091 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17091 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17091 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17091 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17091.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17091.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17091_msk_1.map emd_17091_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17091_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17091_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Small subunit of mitochondrial ribosome in complex with METTL17/Rsm22
| Entire | Name: Small subunit of mitochondrial ribosome in complex with METTL17/Rsm22 |
|---|---|
| Components |
|
-Supramolecule #1: Small subunit of mitochondrial ribosome in complex with METTL17/Rsm22
| Supramolecule | Name: Small subunit of mitochondrial ribosome in complex with METTL17/Rsm22 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#35 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 40 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)