[English] 日本語
 Yorodumi
Yorodumi- EMDB-17087: Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 | |||||||||
 Map data Map data | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Polymerase / uridylation / RNA maturation and turnover control / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of glial cell differentiation / positive regulation of cell proliferation involved in kidney development / negative regulation of pre-miRNA processing / polyuridylation-dependent mRNA catabolic process / uridylyltransferase activity / transposable element silencing by mRNA destabilization / miRNA catabolic process / protein-RNA adaptor activity / RNA uridylyltransferase / RNA 3'-end processing ...negative regulation of glial cell differentiation / positive regulation of cell proliferation involved in kidney development / negative regulation of pre-miRNA processing / polyuridylation-dependent mRNA catabolic process / uridylyltransferase activity / transposable element silencing by mRNA destabilization / miRNA catabolic process / protein-RNA adaptor activity / RNA uridylyltransferase / RNA 3'-end processing / pre-miRNA binding / RNA uridylyltransferase activity / Transcriptional regulation of pluripotent stem cells / Z-decay: degradation of maternal mRNAs by zygotically expressed factors / miRNA metabolic process / Deadenylation of mRNA / pre-miRNA processing / positive regulation of cytoplasmic translation / sequence-specific mRNA binding / oocyte maturation / miRNA binding / stem cell population maintenance / Zygotic genome activation (ZGA) / germ cell development / positive regulation of TOR signaling / rough endoplasmic reticulum / translation initiation factor binding / positive regulation of neuron differentiation / stem cell differentiation / cellular response to glucose stimulus / P-body / cytoplasmic stress granule / G-quadruplex RNA binding / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / negative regulation of translation / mRNA binding / nucleolus / RNA binding / zinc ion binding / nucleoplasm / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
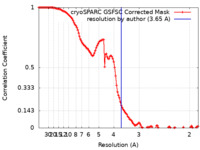
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Yi G / Ye M / Gilbert RJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs. Authors: Gangshun Yi / Mingda Ye / Loic Carrique / Afaf El-Sagheer / Tom Brown / Chris J Norbury / Peijun Zhang / Robert J C Gilbert /  Abstract: Tumor-suppressor let-7 pre-microRNAs (miRNAs) are regulated by terminal uridylyltransferases TUT7 and TUT4 that either promote let-7 maturation by adding a single uridine nucleotide to the pre-miRNA ...Tumor-suppressor let-7 pre-microRNAs (miRNAs) are regulated by terminal uridylyltransferases TUT7 and TUT4 that either promote let-7 maturation by adding a single uridine nucleotide to the pre-miRNA 3' end or mark them for degradation by the addition of multiple uridines. Oligo-uridylation is increased in cells by enhanced TUT7/4 expression and especially by the RNA-binding pluripotency factor LIN28A. Using cryogenic electron microscopy, we captured high-resolution structures of active forms of TUT7 alone, of TUT7 plus pre-miRNA and of both TUT7 and TUT4 bound with pre-miRNA and LIN28A. Our structures reveal that pre-miRNAs engage the enzymes in fundamentally different ways depending on the presence of LIN28A, which clamps them onto the TUTs to enable processive 3' oligo-uridylation. This study reveals the molecular basis for mono- versus oligo-uridylation by TUT7/4, as determined by the presence of LIN28A, and thus their mechanism of action in the regulation of cell fate and in cancer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17087.map.gz emd_17087.map.gz | 151.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17087-v30.xml emd-17087-v30.xml emd-17087.xml emd-17087.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17087_fsc.xml emd_17087_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17087.png emd_17087.png | 100.2 KB | ||
| Filedesc metadata |  emd-17087.cif.gz emd-17087.cif.gz | 7.8 KB | ||
| Others |  emd_17087_half_map_1.map.gz emd_17087_half_map_1.map.gz emd_17087_half_map_2.map.gz emd_17087_half_map_2.map.gz | 151.9 MB 151.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17087 http://ftp.pdbj.org/pub/emdb/structures/EMD-17087 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17087 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17087 | HTTPS FTP |
-Validation report
| Summary document |  emd_17087_validation.pdf.gz emd_17087_validation.pdf.gz | 720.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17087_full_validation.pdf.gz emd_17087_full_validation.pdf.gz | 720.4 KB | Display | |
| Data in XML |  emd_17087_validation.xml.gz emd_17087_validation.xml.gz | 20.1 KB | Display | |
| Data in CIF |  emd_17087_validation.cif.gz emd_17087_validation.cif.gz | 26.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17087 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17087 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17087 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17087 | HTTPS FTP |
-Related structure data
| Related structure data |  8optMC  8oefC  8oppC  8opsC  8ostC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17087.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17087.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 | ||||||||||||||||||||||||||||||||||||
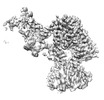
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
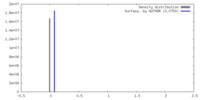
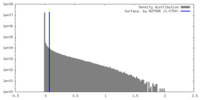
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g...
| File | emd_17087_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 - half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
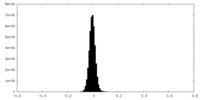
| Density Histograms |
-Half map: Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g...
| File | emd_17087_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 - half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
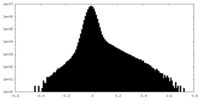
| Density Histograms |
- Sample components
Sample components
-Entire : Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre...
| Entire | Name: Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 |
|---|---|
| Components |
|
-Supramolecule #1: Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre...
| Supramolecule | Name: Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Terminal uridylyltransferase 7 and Protein lin-28 homolog A
| Supramolecule | Name: Terminal uridylyltransferase 7 and Protein lin-28 homolog A type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: RNA (53-MER)
| Supramolecule | Name: RNA (53-MER) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Terminal uridylyltransferase 7
| Macromolecule | Name: Terminal uridylyltransferase 7 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA uridylyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 171.493766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGDTAKPYFV KRTKDRGTMD DDDFRRGHPQ QDYLIIDDHA KGHGSKMEKG LQKKKITPGN YGNTPRKGPC AVSSNPYAFK NPIYSQPAW MNDSHKDQSK RWLSDEHTGN SDNWREFKPG PRIPVINRQR KDSFQENEDG YRWQDTRGCR TVRRLFHKDL T SLETTSEM ...String: MGDTAKPYFV KRTKDRGTMD DDDFRRGHPQ QDYLIIDDHA KGHGSKMEKG LQKKKITPGN YGNTPRKGPC AVSSNPYAFK NPIYSQPAW MNDSHKDQSK RWLSDEHTGN SDNWREFKPG PRIPVINRQR KDSFQENEDG YRWQDTRGCR TVRRLFHKDL T SLETTSEM EAGSPENKKQ RSRPRKPRKT RNEENEQDGD LEGPVIDESV LSTKELLGLQ QAEERLKRDC IDRLKRRPRN YP TAKYTCR LCDVLIESIA FAHKHIKEKR HKKNIKEKQE EELLTTLPPP TPSQINAVGI AIDKVVQEFG LHNENLEQRL EIK RIMENV FQHKLPDCSL RLYGSSCSRL GFKNSDVNID IQFPAIMSQP DVLLLVQECL KNSDSFIDVD ADFHARVPVV VCRE KQSGL LCKVSAGNEN ACLTTKHLTA LGKLEPKLVP LVIAFRYWAK LCSIDRPEEG GLPPYVFALM AIFFLQQRKE PLLPV YLGS WIEGFSLSKL GNFNLQDIEK DVVIWEHTDS AAGDTGITKE EAPRETPIKR GQVSLILDVK HQPSVPVGQL WVELLR FYA LEFNLADLVI SIRVKELVSR ELKDWPKKRI AIEDPYSVKR NVARTLNSQP VFEYILHCLR TTYKYFALPH KITKSSL LK PLNAITCISE HSKEVINHHP DVQTKDDKLK NSVLAQGPGA TSSAANTCKV QPLTLKETAE SFGSPPKEEM GNEHISVH P ENSDCIQADV NSDDYKGDKV YHPETGRKNE KEKVGRKGKH LLTVDQKRGE HVVCGSTRNN ESESTLDLEG FQNPTAKEC EGLATLDNKA DLDGESTEGT EELEDSLNHF THSVQGQTSE MIPSDEEEED DEEEEEEEEP RLTINQREDE DGMANEDELD NTYTGSGDE DALSEEDDEL GEAAKYEDVK ECGKHVERAL LVELNKISLK EENVCEEKNS PVDQSDFFYE FSKLIFTKGK S PTVVCSLC KREGHLKKDC PEDFKRIQLE PLPPLTPKFL NILDQVCIQC YKDFSPTIIE DQAREHIRQN LESFIRQDFP GT KLSLFGS SKNGFGFKQS DLDVCMTING LETAEGLDCV RTIEELARVL RKHSGLRNIL PITTAKVPIV KFFHLRSGLE VDI SLYNTL ALHNTRLLSA YSAIDPRVKY LCYTMKVFTK MCDIGDASRG SLSSYAYTLM VLYFLQQRNP PVIPVLQEIY KGEK KPEIF VDGWNIYFFD QIDELPTYWS ECGKNTESVG QLWLGLLRFY TEEFDFKEHV ISIRRKSLLT TFKKQWTSKY IVIED PFDL NHNLGAGLSR KMTNFIMKAF INGRRVFGIP VKGFPKDYPS KMEYFFDPDV LTEGELAPND RCCRICGKIG HFMKDC PMR RKVRRRRDQE DALNQRYPEN KEKRSKEDKE IHNKYTEREV STKEDKPIQC TPQKAKPMRA AADLGREKIL RPPVEKW KR QDDKDLREKR CFICGREGHI KKECPQFKGS SGSLSSKYMT QGKASAKRTQ QES UniProtKB: Terminal uridylyltransferase 7 |
-Macromolecule #3: Protein lin-28 homolog A
| Macromolecule | Name: Protein lin-28 homolog A / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.778975 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSVSNQQFA GGCAKAAEEA PEEAPEDAAR AADEPQLLHG AGICKWFNVR MGFGFLSMTA RAGVALDPPV DVFVHQSKLH MEGFRSLKE GEAVEFTFKK SAKGLESIRV TGPGGVFCIG SERRPKGKSM QKRRSKGDRC YNCGGLDHHA KECKLPPQPK K CHFCQSIS ...String: MGSVSNQQFA GGCAKAAEEA PEEAPEDAAR AADEPQLLHG AGICKWFNVR MGFGFLSMTA RAGVALDPPV DVFVHQSKLH MEGFRSLKE GEAVEFTFKK SAKGLESIRV TGPGGVFCIG SERRPKGKSM QKRRSKGDRC YNCGGLDHHA KECKLPPQPK K CHFCQSIS HMVASCPLKA QQGPSAQGKP TYFREEEEEI HSPTLLPEAQ N UniProtKB: Protein lin-28 homolog A |
-Macromolecule #2: RNA (53-MER)
| Macromolecule | Name: RNA (53-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.457402 KDa |
| Sequence | String: UUGUACAGUU UGAGGGUAUA UGAUACAACC CGGUACAGGA GAUAACUGUA CAGG |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)