+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Setaria italica NRAT in complex with a nanobody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NRAT / NRAMP / SLC11 / Metal uptake / Aluminium transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaluminum ion transmembrane transporter activity / aluminum cation transport / metal ion transmembrane transporter activity / manganese ion transport / intracellular manganese ion homeostasis / response to aluminum ion / iron ion transmembrane transport / intracellular iron ion homeostasis / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Setaria italica (foxtail millet) / Setaria italica (foxtail millet) /  | |||||||||
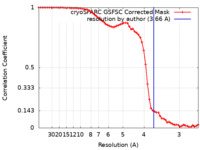
| Method | single particle reconstruction / cryo EM / Resolution: 3.66 Å | |||||||||
 Authors Authors | Ramanadane K / Liziczai M / Markovic D / Straub MS / Rosalen GT / Udovcic A / Dutzler R / Manatschal C | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Structural and functional properties of a plant NRAMP-related aluminum transporter. Authors: Karthik Ramanadane / Márton Liziczai / Dragana Markovic / Monique S Straub / Gian T Rosalen / Anto Udovcic / Raimund Dutzler / Cristina Manatschal /  Abstract: The transport of transition metal ions by members of the SLC11/NRAMP family constitutes a ubiquitous mechanism for the uptake of Fe and Mn across all kingdoms of life. Despite the strong conservation ...The transport of transition metal ions by members of the SLC11/NRAMP family constitutes a ubiquitous mechanism for the uptake of Fe and Mn across all kingdoms of life. Despite the strong conservation of the family, two of its branches have evolved a distinct substrate preference with one mediating Mg uptake in prokaryotes and another the transport of Al into plant cells. Our previous work on the SLC11 transporter from revealed the basis for its Mg selectivity (Ramanadane et al., 2022). Here, we have addressed the structural and functional properties of a putative Al transporter from . We show that the protein transports diverse divalent metal ions and binds the trivalent ions Al and Ga, which are both presumable substrates. Its cryo-electron microscopy (cryo-EM) structure displays an occluded conformation that is closer to an inward- than an outward-facing state, with a binding site that is remodeled to accommodate the increased charge density of its transported substrate. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17000.map.gz emd_17000.map.gz | 11.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17000-v30.xml emd-17000-v30.xml emd-17000.xml emd-17000.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17000_fsc.xml emd_17000_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17000.png emd_17000.png | 105.1 KB | ||
| Masks |  emd_17000_msk_1.map emd_17000_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17000.cif.gz emd-17000.cif.gz | 7 KB | ||
| Others |  emd_17000_half_map_1.map.gz emd_17000_half_map_1.map.gz emd_17000_half_map_2.map.gz emd_17000_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17000 http://ftp.pdbj.org/pub/emdb/structures/EMD-17000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17000 | HTTPS FTP |
-Related structure data
| Related structure data |  8ontMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17000.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17000.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.302 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17000_msk_1.map emd_17000_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17000_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17000_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between SiNRAT and a nanobody used as a fiducial marker f...
| Entire | Name: Complex between SiNRAT and a nanobody used as a fiducial marker for structure determination |
|---|---|
| Components |
|
-Supramolecule #1: Complex between SiNRAT and a nanobody used as a fiducial marker f...
| Supramolecule | Name: Complex between SiNRAT and a nanobody used as a fiducial marker for structure determination type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Setaria italica (foxtail millet) Setaria italica (foxtail millet) |
-Macromolecule #1: NRAMP related aluminium transporter
| Macromolecule | Name: NRAMP related aluminium transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Setaria italica (foxtail millet) Setaria italica (foxtail millet) |
| Molecular weight | Theoretical: 60.147227 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSERAREVGE TGREARHGVL QSGSDTGDHK DKTVELEKDE QFQGQPKWRK FLAHVGPGAL VAIGFLDPSN LETDMQAGAD FKYELLWVV LVGMIFALLI QTLAANLGVK TGRHLAELCR EEYPRYVNIC LWIIAELAVI SDDIPEVLGT AFAFNILLKI P VWAGVILT ...String: MSERAREVGE TGREARHGVL QSGSDTGDHK DKTVELEKDE QFQGQPKWRK FLAHVGPGAL VAIGFLDPSN LETDMQAGAD FKYELLWVV LVGMIFALLI QTLAANLGVK TGRHLAELCR EEYPRYVNIC LWIIAELAVI SDDIPEVLGT AFAFNILLKI P VWAGVILT VFSTLLLLGV QRFGARKLEF IIAAFMFTMA ACFFGELSYL RPSAKEVVKG MFVPSLQGKG AAANAIALFG AI ITPYNLF LHSALVLSRK TPRSVKSIRA ACRYFLIECS LAFIVAFLIN VSVVVVAGTI CNADNLSPTD SNTCSDLTLQ SAP MLLRNV LGRSSSVVYA VALLASGQST TISCTFAGQV IMQGFLDMKM KNWVRNLITR VIAIAPSLIV SIVSGPSGAG KLII FSSMV LSFEMPFALI PLLKFCNSSK KVGPLKESIY TVVIAWILSF ALIVVNTYFL VWTYVDWLVH NHLPKYANAL VSIVV FALM AAYLVFVVYL TFRRDTVSTY VPVSERAQGQ VEAGGAQAVA SAADADQPAP FRKDLADASM ALEVLFQG UniProtKB: Metal transporter |
-Macromolecule #2: Nanobody1
| Macromolecule | Name: Nanobody1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.071546 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QWQLVESGGG LVQAGGSLRL SCVGSGRAFS SGAMGWFRQT PGQEREFVAA ISWSGGSTVY AESVKGRFTI SMDNAKNTVY LRMNSLQPE DTAVYYCAAG TSTFALRRSP EYWGKGTPVT VSS |
-Macromolecule #3: DIUNDECYL PHOSPHATIDYL CHOLINE
| Macromolecule | Name: DIUNDECYL PHOSPHATIDYL CHOLINE / type: ligand / ID: 3 / Number of copies: 1 / Formula: PLC |
|---|---|
| Molecular weight | Theoretical: 622.834 Da |
| Chemical component information |  ChemComp-PLC: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.7 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 69.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)