[English] 日本語
 Yorodumi
Yorodumi- EMDB-16962: Mitochondrial complex I from Mus musculus in the active state bou... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mitochondrial complex I from Mus musculus in the active state bound with piericidin A | |||||||||
 Map data Map data | Full map of mouse complex I inhibited by Piericidin A, post-processed with RELION | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondrial complex I / Respiratory complex I / NADH:ubiquinone oxidoreductase / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial protein import / response to injury involved in regulation of muscle adaptation / Protein lipoylation / Mitochondrial Fatty Acid Beta-Oxidation / Complex I biogenesis / RHOG GTPase cycle / Respiratory electron transport / protein insertion into mitochondrial inner membrane / blastocyst hatching / respiratory system process ...Mitochondrial protein import / response to injury involved in regulation of muscle adaptation / Protein lipoylation / Mitochondrial Fatty Acid Beta-Oxidation / Complex I biogenesis / RHOG GTPase cycle / Respiratory electron transport / protein insertion into mitochondrial inner membrane / blastocyst hatching / respiratory system process / psychomotor behavior / Mitochondrial protein degradation / response to light intensity / cellular response to oxygen levels / mesenchymal stem cell proliferation / iron-sulfur cluster assembly complex / reproductive system development / mitochondrial large ribosomal subunit binding / respiratory chain complex / gliogenesis / mitochondrial [2Fe-2S] assembly complex / mesenchymal stem cell differentiation / circulatory system development / negative regulation of non-canonical NF-kappaB signal transduction / adult walking behavior / positive regulation of mitochondrial membrane potential / response to hydroperoxide / cardiac muscle tissue development / neural precursor cell proliferation / [2Fe-2S] cluster assembly / oxygen sensor activity / cellular response to glucocorticoid stimulus / stem cell division / iron-sulfur cluster assembly / adult behavior / NADH:ubiquinone reductase (H+-translocating) / dopamine metabolic process / mitochondrial ATP synthesis coupled electron transport / positive regulation of ATP biosynthetic process / NADH dehydrogenase activity / proton motive force-driven mitochondrial ATP synthesis / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / positive regulation of execution phase of apoptosis / NADH dehydrogenase (ubiquinone) activity / neuron development / quinone binding / ATP synthesis coupled electron transport / cellular response to interferon-beta / negative regulation of reactive oxygen species biosynthetic process / extrinsic apoptotic signaling pathway / tricarboxylic acid cycle / cellular response to retinoic acid / Neutrophil degranulation / neurogenesis / visual perception / reactive oxygen species metabolic process / muscle contraction / cerebellum development / aerobic respiration / regulation of mitochondrial membrane potential / respiratory electron transport chain / response to nicotine / response to cocaine / mitochondrion organization / DNA damage response, signal transduction by p53 class mediator / kidney development / response to hydrogen peroxide / monooxygenase activity / sensory perception of sound / circadian rhythm / electron transport chain / fatty acid metabolic process / brain development / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / cognition / multicellular organism growth / NAD binding / fatty acid biosynthetic process / positive regulation of protein catabolic process / cellular senescence / FMN binding / nervous system development / myelin sheath / 4 iron, 4 sulfur cluster binding / response to oxidative stress / neuron apoptotic process / gene expression / response to ethanol / in utero embryonic development / response to hypoxia / electron transfer activity / mitochondrial inner membrane / nuclear speck / nuclear body / mitochondrial matrix / response to xenobiotic stimulus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
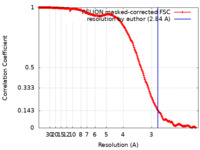
| Method | single particle reconstruction / cryo EM / Resolution: 2.84 Å | |||||||||
 Authors Authors | Grba DN / Chung I / Bridges HR / Agip ANA / Hirst J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I. Authors: Daniel N Grba / Injae Chung / Hannah R Bridges / Ahmed-Noor A Agip / Judy Hirst /  Abstract: Respiratory complex I, a key enzyme in mammalian metabolism, captures the energy released by reduction of ubiquinone by NADH to drive protons across the inner mitochondrial membrane, generating the ...Respiratory complex I, a key enzyme in mammalian metabolism, captures the energy released by reduction of ubiquinone by NADH to drive protons across the inner mitochondrial membrane, generating the proton-motive force for ATP synthesis. Despite remarkable advances in structural knowledge of this complicated membrane-bound enzyme, its mechanism of catalysis remains controversial. In particular, how ubiquinone reduction is coupled to proton pumping and the pathways and mechanisms of proton translocation are contested. We present a 2.4-Å resolution cryo-EM structure of complex I from mouse heart mitochondria in the closed, active (ready-to-go) resting state, with 2945 water molecules modeled. By analyzing the networks of charged and polar residues and water molecules present, we evaluate candidate pathways for proton transfer through the enzyme, for the chemical protons for ubiquinone reduction, and for the protons transported across the membrane. Last, we compare our data to the predictions of extant mechanistic models, and identify key questions to answer in future work to test them. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16962.map.gz emd_16962.map.gz | 457.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16962-v30.xml emd-16962-v30.xml emd-16962.xml emd-16962.xml | 77.2 KB 77.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16962_fsc.xml emd_16962_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_16962.png emd_16962.png | 104.7 KB | ||
| Masks |  emd_16962_msk_1.map emd_16962_msk_1.map | 488.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16962.cif.gz emd-16962.cif.gz | 16.1 KB | ||
| Others |  emd_16962_half_map_1.map.gz emd_16962_half_map_1.map.gz emd_16962_half_map_2.map.gz emd_16962_half_map_2.map.gz | 394.3 MB 394.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16962 http://ftp.pdbj.org/pub/emdb/structures/EMD-16962 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16962 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16962 | HTTPS FTP |
-Related structure data
| Related structure data |  8oltMC  8om1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16962.map.gz / Format: CCP4 / Size: 488.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16962.map.gz / Format: CCP4 / Size: 488.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map of mouse complex I inhibited by Piericidin A, post-processed with RELION | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0625 Å | ||||||||||||||||||||||||||||||||||||
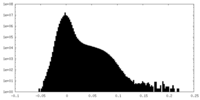
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16962_msk_1.map emd_16962_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
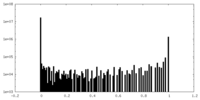
| Density Histograms |
-Half map: Half map 2
| File | emd_16962_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
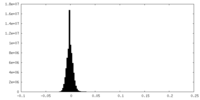
| Density Histograms |
-Half map: Half map 1
| File | emd_16962_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
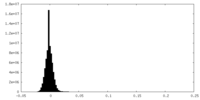
| Density Histograms |
- Sample components
Sample components
+Entire : Mitochondrial respiratory complex I
+Supramolecule #1: Mitochondrial respiratory complex I
+Macromolecule #1: NADH-ubiquinone oxidoreductase chain 3
+Macromolecule #2: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
+Macromolecule #3: NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial
+Macromolecule #4: NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial
+Macromolecule #5: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
+Macromolecule #6: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+Macromolecule #7: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial
+Macromolecule #8: NADH-ubiquinone oxidoreductase chain 1
+Macromolecule #9: NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial
+Macromolecule #10: NADH-ubiquinone oxidoreductase chain 6
+Macromolecule #11: NADH-ubiquinone oxidoreductase chain 4L
+Macromolecule #12: NADH-ubiquinone oxidoreductase chain 5
+Macromolecule #13: NADH-ubiquinone oxidoreductase chain 4
+Macromolecule #14: NADH-ubiquinone oxidoreductase chain 2
+Macromolecule #15: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mi...
+Macromolecule #16: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mit...
+Macromolecule #17: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
+Macromolecule #18: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial
+Macromolecule #19: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
+Macromolecule #20: Acyl carrier protein, mitochondrial
+Macromolecule #21: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5
+Macromolecule #22: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6
+Macromolecule #23: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8
+Macromolecule #24: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11
+Macromolecule #25: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13
+Macromolecule #26: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1
+Macromolecule #27: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3
+Macromolecule #28: NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial
+Macromolecule #29: NADH dehydrogenase [ubiquinone] 1 subunit C2
+Macromolecule #30: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5
+Macromolecule #31: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1
+Macromolecule #32: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mit...
+Macromolecule #33: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mito...
+Macromolecule #34: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6
+Macromolecule #35: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, mito...
+Macromolecule #36: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3
+Macromolecule #37: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mito...
+Macromolecule #38: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4
+Macromolecule #39: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9
+Macromolecule #40: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7, METH...
+Macromolecule #41: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10
+Macromolecule #42: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+Macromolecule #43: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7
+Macromolecule #44: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial
+Macromolecule #45: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #46: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #47: IRON/SULFUR CLUSTER
+Macromolecule #48: Piericidin A
+Macromolecule #49: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #50: FLAVIN MONONUCLEOTIDE
+Macromolecule #51: SODIUM ION
+Macromolecule #52: CARDIOLIPIN
+Macromolecule #53: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #54: MAGNESIUM ION
+Macromolecule #55: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #56: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #57: ZINC ION
+Macromolecule #58: ~{S}-[2-[3-[[(2~{R})-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butan...
+Macromolecule #59: MYRISTIC ACID
+Macromolecule #60: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.14 Component:
Details: PH corrected at room temperature | ||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR Details: The grid was treated for 48 hours in an anaerobic glovebox in ethanol containing 5mM 11-mercaptoundecylhexaethyleneglycol, washed three times in ethanol and air dried prior to use. Note the ...Details: The grid was treated for 48 hours in an anaerobic glovebox in ethanol containing 5mM 11-mercaptoundecylhexaethyleneglycol, washed three times in ethanol and air dried prior to use. Note the hole sizes are R0.6/1. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 10 to 12 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: COUNTING / #0 - Digitization - Dimensions - Width: 3838 pixel / #0 - Digitization - Dimensions - Height: 3710 pixel / #0 - Digitization - Frames/image: 1-21 / #0 - Number grids imaged: 1 / #0 - Number real images: 1200 / #0 - Average exposure time: 10.0 sec. / #0 - Average electron dose: 50.0 e/Å2 #0 - Details: Images were collected over 25 frames per exposure #1 - Image recording ID: 2 / #1 - Film or detector model: FEI FALCON III (4k x 4k) / #1 - Detector mode: COUNTING / #1 - Number grids imaged: 1 / #1 - Number real images: 1454 / #1 - Average exposure time: 71.5 sec. / #1 - Average electron dose: 46.0 e/Å2 #1 - Details: Images were collected over 40 frames per exposure |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.4 µm / Nominal defocus min: 2.2 µm / Nominal magnification: 47600 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)