+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | S.cerevisiae THO complex from endogenous nuclear mRNPs | ||||||||||||
 マップデータ マップデータ | dimer masked map | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | Complex / Nuclear / Dimer / NUCLEAR PROTEIN | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報nucleoplasmic THO complex / cellular response to azide / THO complex / THO complex part of transcription export complex / positive regulation of transcription elongation by RNA polymerase I / transcription export complex / Cdc73/Paf1 complex / mRNA 3'-end processing / positive regulation of transcription by RNA polymerase I / mRNA export from nucleus ...nucleoplasmic THO complex / cellular response to azide / THO complex / THO complex part of transcription export complex / positive regulation of transcription elongation by RNA polymerase I / transcription export complex / Cdc73/Paf1 complex / mRNA 3'-end processing / positive regulation of transcription by RNA polymerase I / mRNA export from nucleus / transcription-coupled nucleotide-excision repair / stress granule assembly / transcription elongation by RNA polymerase II / mRNA processing / DNA recombination / nucleic acid binding / molecular adaptor activity / chromosome, telomeric region / mRNA binding / nucleus 類似検索 - 分子機能 | ||||||||||||
| 生物種 |   | ||||||||||||
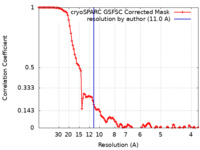
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 11.0 Å | ||||||||||||
 データ登録者 データ登録者 | Bonneau F / Schaefer IB / Conti E | ||||||||||||
| 資金援助 | European Union,  ドイツ, ドイツ,  デンマーク, 3件 デンマーク, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Genes Dev / 年: 2023 ジャーナル: Genes Dev / 年: 2023タイトル: Nuclear mRNPs are compact particles packaged with a network of proteins promoting RNA-RNA interactions. 著者: Fabien Bonneau / Jérôme Basquin / Barbara Steigenberger / Tillman Schäfer / Ingmar B Schäfer / Elena Conti /  要旨: Messenger RNAs (mRNAs) are at the center of the central dogma of molecular biology. In eukaryotic cells, these long ribonucleic acid polymers do not exist as naked transcripts; rather, they associate ...Messenger RNAs (mRNAs) are at the center of the central dogma of molecular biology. In eukaryotic cells, these long ribonucleic acid polymers do not exist as naked transcripts; rather, they associate with mRNA-binding proteins to form messenger ribonucleoprotein (mRNP) complexes. Recently, global proteomic and transcriptomic studies have provided comprehensive inventories of mRNP components. However, knowledge of the molecular features of distinct mRNP populations has remained elusive. We purified endogenous nuclear mRNPs from by harnessing the mRNP biogenesis factors THO and Sub2 in biochemical procedures optimized to preserve the integrity of these transient ribonucleoprotein assemblies. We found that these mRNPs are compact particles that contain multiple copies of Yra1, an essential protein with RNA-annealing properties. To investigate their molecular and architectural organization, we used a combination of proteomics, RNA sequencing, cryo-electron microscopy, cross-linking mass spectrometry, structural models, and biochemical assays. Our findings indicate that yeast nuclear mRNPs are packaged around an intricate network of interconnected proteins capable of promoting RNA-RNA interactions via their positively charged intrinsically disordered regions. The evolutionary conservation of the major mRNA-packaging factor (yeast Yra1 and Aly/REF in metazoans) points toward a general paradigm governing nuclear mRNP packaging. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_16841.map.gz emd_16841.map.gz | 121.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-16841-v30.xml emd-16841-v30.xml emd-16841.xml emd-16841.xml | 22.9 KB 22.9 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_16841_fsc.xml emd_16841_fsc.xml | 10.8 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_16841.png emd_16841.png | 27.4 KB | ||
| マスクデータ |  emd_16841_msk_1.map emd_16841_msk_1.map | 129.7 MB |  マスクマップ マスクマップ | |
| その他 |  emd_16841_half_map_1.map.gz emd_16841_half_map_1.map.gz emd_16841_half_map_2.map.gz emd_16841_half_map_2.map.gz | 120.3 MB 120.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16841 http://ftp.pdbj.org/pub/emdb/structures/EMD-16841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16841 | HTTPS FTP |
-関連構造データ
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
|---|
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_16841.map.gz / 形式: CCP4 / 大きさ: 129.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_16841.map.gz / 形式: CCP4 / 大きさ: 129.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | dimer masked map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.885 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_16841_msk_1.map emd_16841_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
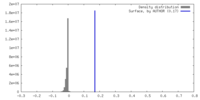
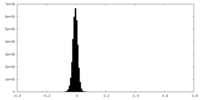
| 密度ヒストグラム |
-ハーフマップ: dimer half map A
| ファイル | emd_16841_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | dimer half map A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
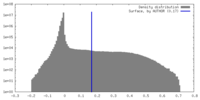
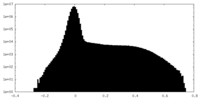
| 密度ヒストグラム |
-ハーフマップ: dimer half map B
| ファイル | emd_16841_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | dimer half map B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
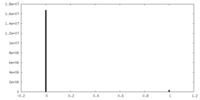
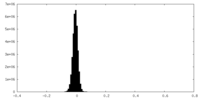
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : THO complex dimer
| 全体 | 名称: THO complex dimer |
|---|---|
| 要素 |
|
-超分子 #1: THO complex dimer
| 超分子 | 名称: THO complex dimer / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all 詳細: From nuclear mRNPs isolated by bi-molecular affinity purification using Sub2 and Hpr1 as baits, then treated with Benzonase. |
|---|---|
| 由来(天然) | 生物種:  |
-分子 #1: THO complex subunit 2
| 分子 | 名称: THO complex subunit 2 / タイプ: protein_or_peptide / ID: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 配列 | 文字列: MAEQTLLSKL NALSQKVIPP ASPSQASILT EEVIRNWPER SKTLCSDFTA LESNDEKEDW LRTLFIELFD FINKNDENSP LKLSDVASFT NELVNHERQV SQASIVGKMF IAVSSTVPNI NDLTTISLCK LIPSLHEELF KFSWISSKLL NKEQTTLLRH LLKKSKYELK ...文字列: MAEQTLLSKL NALSQKVIPP ASPSQASILT EEVIRNWPER SKTLCSDFTA LESNDEKEDW LRTLFIELFD FINKNDENSP LKLSDVASFT NELVNHERQV SQASIVGKMF IAVSSTVPNI NDLTTISLCK LIPSLHEELF KFSWISSKLL NKEQTTLLRH LLKKSKYELK KYNLLVENSV GYGQLVALLI LAYYDPDNFS KVSAYLKEIY HIMGKYSLDS IRTLDVILNV SSQFITEGYK FFIALLRKSD SWPSSHVANN SNYSSLNEGG NMIAANIISF NLSQYNEEVD KENYERYMDM CCILLKNGFV NFYSIWDNVK PEMEFLQEYI QNLETELEEE STKGVENPLA MAAALSTENE TDEDNALVVN DDVNMKDKIS EETNADIESK GKQKTQQDIL LFGKIKLLER LLIHGCVIPV IHVLKQYPKV LYVSESLSRY LGRVFEYLLN PLYTSMTSSG ESKDMATALM ITRIDNGILA HKPRLIHKYK THEPFESLEL NSSYVFYYSE WNSNLTPFAS VNDLFENSHI YLSIIGPYLG RIPTLLSKIS RIGVADIQKN HGSESLHVTI DKWIDYVRKF IFPATSLLQN NPIATSEVYE LMKFFPFEKR YFIYNEMMTK LSQDILPLKV SFNKAEREAK SILKALSIDT IAKESRRFAK LISTNPLASL VPAVKQIENY DKVSELVVYT TKYFNDFAYD VLQFVLLLRL TYNRPAVQFD GVNQAMWVQR LSIFIAGLAK NCPNMDISNI ITYILKTLHN GNIIAVSILK ELIITVGGIR DLNEVNMKQL LMLNSGSPLK QYARHLIYDF RDDNSVISSR LTSFFTDQSA ISEIILLLYT LNLKANTQNS HYKILSTRCD EMNTLLWSFI ELIKHCLKGK AFEENVLPFV ELNNRFHLST PWTFHIWRDY LDNQLNSNEN FSIDELIEGA EFSDVDLTKI SKDLFTTFWR LSLYDIHFDK SLYDERKNAL SGENTGHMSN RKKHLIQNQI KDILVTGISH QRAFKKTSEF ISEKSNVWNK DCGEDQIKIF LQNCVVPRVL FSPSDALFSS FFIFMAFRTE NLMSILNTCI TSNILKTLLF CCTSSEAGNL GLFFTDVLKK LEKMRLNGDF NDQASRKLYE WHSVITEQVI DLLSEKNYMS IRNGIEFMKH VTSVFPVVKA HIQLVYTTLE ENLINEERED IKLPSSALIG HLKARLKDAL ELDEFCTLTE EEAEQKRIRE MELEEIKNYE TACQNEQKQV ALRKQLELNK SQRLQNDPPK SVASGSAGLN SKDRYTYSRN EPVIPTKPSS SQWSYSKVTR HVDDINHYLA TNHLQKAISL VENDDETRNL RKLSKQNMPI FDFRNSTLEI FERYFRTLIQ NPQNPDFAEK IDSLKRYIKN ISREPYPDTT SSYSEAAAPE YTKRSSRYSG NAGGKDGYGS SNYRGPSNDR SAPKNIKPIS SYAHKRSELP TRPSKSKTYN DRSRALRPTG PDRGDGFDQR DNRLREEYKK NSSQRSQLRF PEKPFQEGKD SSKANPYQAS SYKRDSPSEN EEKPNKRFKK DETIRNKFQT QDYRNTRDSG AAHRANENQR YNGNRKSNTQ ALPQGPKGGN YVSRYQR UniProtKB: THO complex subunit 2 |
-分子 #2: THO complex subunit HPR1
| 分子 | 名称: THO complex subunit HPR1 / タイプ: protein_or_peptide / ID: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 配列 | 文字列: MSNTEELIQN SIGFLQKTFK ALPVSFDSIR HEPLPSSMLH ASVLNFEWEP LEKNISAIHD RDSLIDIILK RFIIDSMTNA IEDEEENNLE KGLLNSCIGL DFVYNSRFNR SNPASWGNTF FELFSTIIDL LNSPSTFLKF WPYAESRIEW FKMNTSVEPV SLGESNLISY ...文字列: MSNTEELIQN SIGFLQKTFK ALPVSFDSIR HEPLPSSMLH ASVLNFEWEP LEKNISAIHD RDSLIDIILK RFIIDSMTNA IEDEEENNLE KGLLNSCIGL DFVYNSRFNR SNPASWGNTF FELFSTIIDL LNSPSTFLKF WPYAESRIEW FKMNTSVEPV SLGESNLISY KQPLYEKLRH WNDILAKLEN NDILNTVKHY NMKYKLENFL SELLPINEES NFNRSASISA LQESDNEWNR SARERESNRS SDVIFAADYN FVFYHLIICP IEFAFSDLEY KNDVDRSLSP LLDAILEIEE NFYSKIKMNN RTRYSLEEAL NTEYYANYDV MTPKLPVYMK HSNAMKMDRN EFWANLQNIK ESDDYTLRPT IMDISLSNTT CLYKQLTQED DDYYRKQFIL QLCFTTNLIR NLISSDETRN FYKSCYLREN PLSDIDFENL DEVNKKRGLN LCSYICDNRV LKFYKIKDPD FYRVIRKLMS SDEKFTTAKI DGFKEFQNFR ISKEKIPPPA FDETFKKFTF IKMGNKLINN VWKIPTGLDK IEQEVKKPEG VYEAAQAKWE SKISSETSGG EAKDEIIRQW QTLRFLRSRY LFDFDKVNEK TGVDGLFEEP RKVEALDDSF KEKLLYKINQ EHRKKLQDAR EYKIGKERKK RALEEEASFP EREQKIKSQR INSASQTEGD ELKSEQTQPK GEISEENTKI KSSEVSSQDP DSGVAGEFAP QNTTAQLENP KTEDNNAATS NISNGSSTQD MKGSGSGSGS GSSAWSHPQF EKGGGSGGGS GGSAWSHPQF EK UniProtKB: THO complex subunit HPR1 |
-分子 #3: THO complex subunit MFT1
| 分子 | 名称: THO complex subunit MFT1 / タイプ: protein_or_peptide / ID: 3 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 配列 | 文字列: MPLSQKQIDQ VRTKVHYSEV DTPFNKYLDI LGKVTKLTGS IINGTLSNDD SKIEKLTEQN ISQLKESAHL RFLDLQSSID TKKVADENWE TCQQETLAKL ENLKDKLPDI KSIHSKLLLR IGKLQGLYDS VQVINREVEG LSEGRTSLVV TRAEWEKELG TDLVKFLIEK ...文字列: MPLSQKQIDQ VRTKVHYSEV DTPFNKYLDI LGKVTKLTGS IINGTLSNDD SKIEKLTEQN ISQLKESAHL RFLDLQSSID TKKVADENWE TCQQETLAKL ENLKDKLPDI KSIHSKLLLR IGKLQGLYDS VQVINREVEG LSEGRTSLVV TRAEWEKELG TDLVKFLIEK NYLKLVDPGL KKDSSEERYR IYDDFSKGPK ELESINASMK SDIENVRQEV SSYKEKWLRD AEIFGKITSI FKEELLKRDG LLNEAEGDNI DEDYESDEDE ERKERFKRQR SMVEVNTIEN VDEKEESDHE YDDQEDEENE EEDDMEVDVE DIKEDNEVDG ESSQQEDNSR QGNNEETDKE TGVIEEPDAV NDAEEADSDH SSRKLGGTTS DFSASSSVEE VK UniProtKB: THO complex subunit MFT1 |
-分子 #4: Protein TEX1
| 分子 | 名称: Protein TEX1 / タイプ: protein_or_peptide / ID: 4 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 配列 | 文字列: MSTIGAVDIL NQKTITSEVA ASVTSKYLQS TFSKGNTSHI EDKRFIHVSS RSHSRFTSTP ITPNEILSLK FHVSGSSMAY SRMDGSLTVW FIKDASFDKS VEVYIPDCCG SDKLATDLSW NPTSLNQIAV VSNSSEISLL LINEKSLTAS KLRTLSLGSK TKVNTCLYDP ...文字列: MSTIGAVDIL NQKTITSEVA ASVTSKYLQS TFSKGNTSHI EDKRFIHVSS RSHSRFTSTP ITPNEILSLK FHVSGSSMAY SRMDGSLTVW FIKDASFDKS VEVYIPDCCG SDKLATDLSW NPTSLNQIAV VSNSSEISLL LINEKSLTAS KLRTLSLGSK TKVNTCLYDP LGNWLLAATK SEKIYLFDVK KDHSSVCSLN ISDISQEDND VVYSLAWSNG GSHIFIGFKS GYLAILKAKH GILEVCTKIK AHTGPITEIK MDPWGRNFIT GSIDGNCYVW NMKSLCCELI INDLNSAVTT LDVCHLGKIL GICTEDEMVY FYDLNSGNLL HSKSLANYKT DPVLKFYPDK SWYIMSGKND TLSNHFVKNE KNLITYWKDM FDNTMIEKRR KNNGGGNNHN KRTSKNTDRI GKDRPSRFNS KK UniProtKB: Protein TEX1 |
-分子 #5: THO complex subunit THP2
| 分子 | 名称: THO complex subunit THP2 / タイプ: protein_or_peptide / ID: 5 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 配列 | 文字列: MTKEEGRTYF ESLCEEEQSL QESQTHLLNI LDILSVLADP RSSDDLLTES LKKLPDLHRE LINSSIRLRY DKYQTREAQL LEDTKTGRDV AAGVQNPKSI SEYYSTFEHL NRDTLRYINL LKRLSVDLAK QVEVSDPSVT VYEMDKWVPS EKLQGILEQY CAPDTDIRGV ...文字列: MTKEEGRTYF ESLCEEEQSL QESQTHLLNI LDILSVLADP RSSDDLLTES LKKLPDLHRE LINSSIRLRY DKYQTREAQL LEDTKTGRDV AAGVQNPKSI SEYYSTFEHL NRDTLRYINL LKRLSVDLAK QVEVSDPSVT VYEMDKWVPS EKLQGILEQY CAPDTDIRGV DAQIKNYLDQ IKMARAKFGL ENKYSLKERL STLTKELNHW RKEWDDIEML MFGDDAHSMK KMIQKIDSLK SEINAPSESY PVDKEGDIVL E UniProtKB: THO complex subunit THP2 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 8 構成要素:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| グリッド | モデル: Quantifoil R2/1 / 材質: COPPER / メッシュ: 200 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 20 sec. / 前処理 - 雰囲気: AIR | ||||||||
| 凍結 | 凍結剤: ETHANE-PROPANE / チャンバー内湿度: 95 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS GLACIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - 画像ごとのフレーム数: 1-40 / 撮影したグリッド数: 1 / 実像数: 2258 / 平均露光時間: 16.0 sec. / 平均電子線量: 59.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.62 mm / 最大 デフォーカス(公称値): 3.0 µm / 最小 デフォーカス(公称値): 1.5 µm / 倍率(公称値): 22000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
 ムービー
ムービー コントローラー
コントローラー




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)