+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RcpA-TadD local refinement map | |||||||||
 Map data Map data | RcpA-TadD Local refinement map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RcpA / TadD / Secretin / Pilotin / Tight adherence secretion system / local refinement / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtype II protein secretion system complex / protein secretion / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Tassinari M / Low HH | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Assembly mechanism of a Tad secretion system secretin-pilotin complex. Authors: Matteo Tassinari / Marta Rudzite / Alain Filloux / Harry H Low /    Abstract: The bacterial Tight adherence Secretion System (TadSS) assembles surface pili that drive cell adherence, biofilm formation and bacterial predation. The structure and mechanism of the TadSS is mostly ...The bacterial Tight adherence Secretion System (TadSS) assembles surface pili that drive cell adherence, biofilm formation and bacterial predation. The structure and mechanism of the TadSS is mostly unknown. This includes characterisation of the outer membrane secretin through which the pilus is channelled and recruitment of its pilotin. Here we investigate RcpA and TadD lipoprotein from Pseudomonas aeruginosa. Light microscopy reveals RcpA colocalising with TadD in P. aeruginosa and when heterologously expressed in Escherichia coli. We use cryogenic electron microscopy to determine how RcpA and TadD assemble a secretin channel with C13 and C14 symmetries. Despite low sequence homology, we show that TadD shares a similar fold to the type 4 pilus system pilotin PilF. We establish that the C-terminal four residues of RcpA bind TadD - an interaction essential for secretin formation. The binding mechanism between RcpA and TadD appears distinct from known secretin-pilotin pairings in other secretion systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16818.map.gz emd_16818.map.gz | 7.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16818-v30.xml emd-16818-v30.xml emd-16818.xml emd-16818.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16818_fsc.xml emd_16818_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16818.png emd_16818.png | 47.2 KB | ||
| Others |  emd_16818_half_map_1.map.gz emd_16818_half_map_1.map.gz emd_16818_half_map_2.map.gz emd_16818_half_map_2.map.gz | 45.7 MB 45.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16818 http://ftp.pdbj.org/pub/emdb/structures/EMD-16818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16818 | HTTPS FTP |
-Validation report
| Summary document |  emd_16818_validation.pdf.gz emd_16818_validation.pdf.gz | 692.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16818_full_validation.pdf.gz emd_16818_full_validation.pdf.gz | 691.9 KB | Display | |
| Data in XML |  emd_16818_validation.xml.gz emd_16818_validation.xml.gz | 14.2 KB | Display | |
| Data in CIF |  emd_16818_validation.cif.gz emd_16818_validation.cif.gz | 19.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16818 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16818 | HTTPS FTP |
-Related structure data
| Related structure data |  8odnC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16818.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16818.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RcpA-TadD Local refinement map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
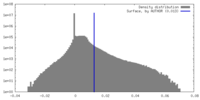
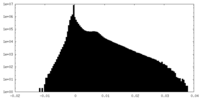
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: RcpA-TadD Local refinement map
| File | emd_16818_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RcpA-TadD Local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
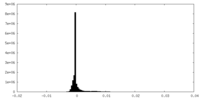
| Density Histograms |
-Half map: RcpA-TadD Local refinement map
| File | emd_16818_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RcpA-TadD Local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
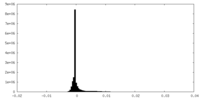
| Density Histograms |
- Sample components
Sample components
-Entire : RcpA-TadD from the Paseudomonas aeruginosa Tad secretion system
| Entire | Name: RcpA-TadD from the Paseudomonas aeruginosa Tad secretion system |
|---|---|
| Components |
|
-Supramolecule #1: RcpA-TadD from the Paseudomonas aeruginosa Tad secretion system
| Supramolecule | Name: RcpA-TadD from the Paseudomonas aeruginosa Tad secretion system type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 900 KDa |
-Macromolecule #1: RcpA
| Macromolecule | Name: RcpA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MHRSTGIGVS RWLGGLLGVA LALPALALPQ GCIELLAQAP RVDVVQGQQR DLRLAVPIER LAIGDPKIAD VQLLDRRGFL VTGKEQGSTS LLIWTGCSPE PLRSLVEVEG RGSVDTRGAP AFTVGAAEEL PNQVQTDIRF VEVSRSKLKQ ASTSFVRRGG NLWVLGAPGS ...String: MHRSTGIGVS RWLGGLLGVA LALPALALPQ GCIELLAQAP RVDVVQGQQR DLRLAVPIER LAIGDPKIAD VQLLDRRGFL VTGKEQGSTS LLIWTGCSPE PLRSLVEVEG RGSVDTRGAP AFTVGAAEEL PNQVQTDIRF VEVSRSKLKQ ASTSFVRRGG NLWVLGAPGS LGDIKVNADG SGLGGTFGTG SSGFNLIFGG GKWLSFMNAL EGSGFAYTLA RPSLVAMSGQ SASFLAGGEF PIPVPNGTND NVTIEYKEFG IRLTLTPTVM NNRRIALKVA PEVSELDYSA GIQSGGVAVP ALRVRRTDTS VMLADGESFV ISGLTSSNSV SNVDKFPWLG DIPILGAFFR STKLDKDDRE LLMIVTPHLV QPLAADAQLP DLPGEGLRHY DPGFSRLYFL ERGEYDGQQN DTGLSDSAWS HPQFEK UniProtKB: RcpA |
-Macromolecule #2: TadD
| Macromolecule | Name: TadD / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MKALIGIGLC AALLGGCAAL PGRDGPRECS QQLGQEQELQ MNMVRDMIRE GRLHAALANL ESMPPGLLDV REERALILRR IGDPRARAEY QALLETCKAP EAHHGLGLLA LRNGDSARAV LELREAARLR PTESRFRNDL GVALLKRGDR VGARFEFITA LELQQGGKLP ...String: MKALIGIGLC AALLGGCAAL PGRDGPRECS QQLGQEQELQ MNMVRDMIRE GRLHAALANL ESMPPGLLDV REERALILRR IGDPRARAEY QALLETCKAP EAHHGLGLLA LRNGDSARAV LELREAARLR PTESRFRNDL GVALLKRGDR VGARFEFITA LELQQGGKLP ATNLLGLLYL QGDREDAQRL IERLQLDARD IRAAEARARS WGAVPTPDAA PASDDPLAEL PAEANMHTAM ANEAPGSDYK DDDDK UniProtKB: TPR repeat-containing protein PA4299 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: 50 mM Hepes pH 8, 100 mM NaCl, 2 mM EDTA, 0.06 % bDDM | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)