+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human MUC5B amino acids 26-1435 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mucin / Filament / D assemblies / disulfide bonds / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective GALNT3 causes HFTC / Defective C1GALT1C1 causes TNPS / Defective GALNT12 causes CRCS1 / Termination of O-glycan biosynthesis / O-linked glycosylation of mucins / Dectin-2 family / extracellular matrix / Golgi lumen / intracellular membrane-bounded organelle / extracellular space ...Defective GALNT3 causes HFTC / Defective C1GALT1C1 causes TNPS / Defective GALNT12 causes CRCS1 / Termination of O-glycan biosynthesis / O-linked glycosylation of mucins / Dectin-2 family / extracellular matrix / Golgi lumen / intracellular membrane-bounded organelle / extracellular space / extracellular exosome / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Khmelnitsky L / Javitt G / Elad N / Fass D | |||||||||
| Funding support |  Israel, 1 items Israel, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Pulmonary Fibrosis Mutation impairs Mucin 5B Supramolecular Assembly Authors: Khmelnitsky L / Javitt G / Reznik N / Yeshaya N / Elad N / Anikster Y / Shalva N / Barel O / Pode-Shakked B / Greenberg LJ / Segel MJ / Fass D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16808.map.gz emd_16808.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16808-v30.xml emd-16808-v30.xml emd-16808.xml emd-16808.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16808.png emd_16808.png | 63.1 KB | ||
| Filedesc metadata |  emd-16808.cif.gz emd-16808.cif.gz | 5.4 KB | ||
| Others |  emd_16808_half_map_1.map.gz emd_16808_half_map_1.map.gz emd_16808_half_map_2.map.gz emd_16808_half_map_2.map.gz | 1.2 GB 1.2 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16808 http://ftp.pdbj.org/pub/emdb/structures/EMD-16808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16808 | HTTPS FTP |
-Related structure data
| Related structure data |  8oerM  8oesM M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16808.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16808.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8264 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16808_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
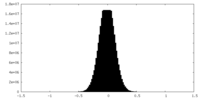
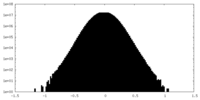
| Density Histograms |
-Half map: #2
| File | emd_16808_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
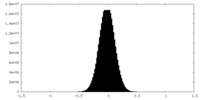
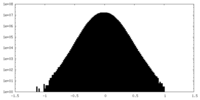
| Density Histograms |
- Sample components
Sample components
-Entire : MUC5B amino acids 26-1435
| Entire | Name: MUC5B amino acids 26-1435 |
|---|---|
| Components |
|
-Supramolecule #1: MUC5B amino acids 26-1435
| Supramolecule | Name: MUC5B amino acids 26-1435 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: MUC5B
| Macromolecule | Name: MUC5B / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QGPVEPSWEN AGHTMDGGAP TSSPTRRVSF VPPVTVFPSL SPLNPAHNGR VCSTWGDFHY KTFDGDVFRF PGLCNYVFSE HCRAAYEDFN VQLRRGLVGS RPVVTRVVIK AQGLVLEASN GSVLINGQRE ELPYSRTGLL VEQSGDYIKV SIRLVLTFLW NGEDSALLEL ...String: QGPVEPSWEN AGHTMDGGAP TSSPTRRVSF VPPVTVFPSL SPLNPAHNGR VCSTWGDFHY KTFDGDVFRF PGLCNYVFSE HCRAAYEDFN VQLRRGLVGS RPVVTRVVIK AQGLVLEASN GSVLINGQRE ELPYSRTGLL VEQSGDYIKV SIRLVLTFLW NGEDSALLEL DPKYANQTCG LCGDFNGLPA FNEFYAHNAR LTPLQFGNLQ KLDGPTEQCP DPLPLPAGNC TDEEGICHRT LLGPAFAECH ALVDSTAYLA ACAQDLCRCP TCPCATFVEY SRQCAHAGGQ PRNWRCPELC PRTCPLNMQH QECGSPCTDT CSNPQRAQLC EDHCVDGCFC PPGTVLDDIT HSGCLPLGQC PCTHGGRTYS PGTSFNTTCS SCTCSGGLWQ CQDLPCPGTC SVQGGAHIST YDEKLYDLHG DCSYVLSKKC ADSSFTVLAE LRKCGLTDNE NCLKAVTLSL DGGDTAIRVQ ADGGVFLNSI YTQLPLSAAN ITLFTPSSFF IVVQTGLGLQ LLVQLVPLMQ VFVRLDPAHQ GQMCGLCGNF NQNQADDFTA LSGVVEATGA AFANTWKAQA ACANARNSFE DPCSLSVENE NYARHWCSRL TDPNSAFSRC HSIINPKPFH SNCMFDTCNC ERSEDCLCAA LSSYVHACAA KGVQLSDWRD GVCTKYMQNC PKSQRYAYVV DACQPTCRGL SEADVTCSVS FVPVDGCTCP AGTFLNDAGA CVPAQECPCY AHGTVLAPGE VVHDEGAVCS CTGGKLSCLG ASLQKSTGCA APMVYLDCSN SSAGTPGAEC LRSCHTLDVG CFSTHCVSGC VCPPGLVSDG SGGCIAEEDC PCVHNEATYK PGETIRVDCN TCTCRNRRWE CSHRLCLGTC VAYGDGHFIT FDGDRYSFEG SCEYILAQDY CGDNTTHGTF RIVTENIPCG TTGTTCSKAI KLFVESYELI LQEGTFKAVA RGPGGDPPYK IRYMGIFLVI ETHGMAVSWD RKTSVFIRLH QDYKGRVCGL CGNFDDNAIN DFATRSRSVV GDALEFGNSW KLSPSCPDAL APKDPCTANP FRKSWAQKQC SILHGPTFAA CRSQVDSTKY YEACVNDACA CDSGGDCECF CTAVAAYAQA CHDAGLCVSW RTPDTCPLFC DFYNPHGGCE WHYQPCGAPC LKTCRNPSGH CLVDLPGLEG CYPKCPPSQP FFNEDQMKCV AQCGCYDKDG NYYDVGARVP TAENCQSCNC TPSGIQCAHS LEACTCTYED RTYSYQDVIY NTTDGLGACL IAICGSNGTI IRKAVACPGT PATTPFTFTT AWVPHSTTSP ALPVSTVCVR EVCRWSSWYN GHRPEPGLGG GDFETFENLR QRGYQVCPVL ADIECRAAQL PDMPLEELGQ QVDCDRMRGL MCANSQQSPP LCHDYELRVL CCEYVPCGPS HHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.45 mg/mL |
|---|---|
| Buffer | pH: 5.2 / Component - Concentration: 100.0 mM / Component - Formula: C6H13NO4S / Component - Name: MES |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 88708 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)