[English] 日本語
 Yorodumi
Yorodumi- EMDB-16777: Structure of the Nucleosome Core Particle from Trypanosoma brucei -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Nucleosome Core Particle from Trypanosoma brucei | |||||||||
 Map data Map data | Masked sharpened map of Nucleosome core particle from Trypanosoma brucei | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleosome Chromatin Parasite Trypanosome Kinetoplast / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnuclear lumen / ciliary plasm / ciliary transition zone / phosphate ion binding / chromosome organization / structural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / protein heterodimerization activity ...nuclear lumen / ciliary plasm / ciliary transition zone / phosphate ion binding / chromosome organization / structural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / protein heterodimerization activity / DNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Sandoval G / Deak G / Tuijtel MW / Wilson MD | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Histone divergence in trypanosomes results in unique alterations to nucleosome structure. Authors: Gauri Deák / Hannah Wapenaar / Gorka Sandoval / Ruofan Chen / Mark R D Taylor / Hayden Burdett / James A Watson / Maarten W Tuijtel / Shaun Webb / Marcus D Wilson /   Abstract: Eukaryotes have a multitude of diverse mechanisms for organising and using their genomes, but the histones that make up chromatin are highly conserved. Unusually, histones from kinetoplastids are ...Eukaryotes have a multitude of diverse mechanisms for organising and using their genomes, but the histones that make up chromatin are highly conserved. Unusually, histones from kinetoplastids are highly divergent. The structural and functional consequences of this variation are unknown. Here, we have biochemically and structurally characterised nucleosome core particles (NCPs) from the kinetoplastid parasite Trypanosoma brucei. A structure of the T. brucei NCP reveals that global histone architecture is conserved, but specific sequence alterations lead to distinct DNA and protein interaction interfaces. The T. brucei NCP is unstable and has weakened overall DNA binding. However, dramatic changes at the H2A-H2B interface introduce local reinforcement of DNA contacts. The T. brucei acidic patch has altered topology and is refractory to known binders, indicating that the nature of chromatin interactions in T. brucei may be unique. Overall, our results provide a detailed molecular basis for understanding evolutionary divergence in chromatin structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16777.map.gz emd_16777.map.gz | 113.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16777-v30.xml emd-16777-v30.xml emd-16777.xml emd-16777.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16777.png emd_16777.png | 64.1 KB | ||
| Masks |  emd_16777_msk_1.map emd_16777_msk_1.map | 120.4 MB |  Mask map Mask map | |
| Others |  emd_16777_additional_1.map.gz emd_16777_additional_1.map.gz emd_16777_half_map_1.map.gz emd_16777_half_map_1.map.gz emd_16777_half_map_2.map.gz emd_16777_half_map_2.map.gz | 59.9 MB 111.6 MB 111.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16777 http://ftp.pdbj.org/pub/emdb/structures/EMD-16777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16777 | HTTPS FTP |
-Related structure data
| Related structure data |  8comMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16777.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16777.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked sharpened map of Nucleosome core particle from Trypanosoma brucei | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.829 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16777_msk_1.map emd_16777_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
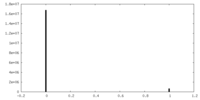
| Density Histograms |
-Additional map: unsharpened map of Nucleosome core particle from Trypanosoma brucei
| File | emd_16777_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map of Nucleosome core particle from Trypanosoma brucei | ||||||||||||
| Projections & Slices |
| ||||||||||||
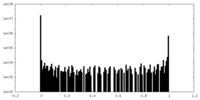
| Density Histograms |
-Half map: half map 2 of Nucleosome core particle from Trypanosoma brucei
| File | emd_16777_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 of Nucleosome core particle from Trypanosoma brucei | ||||||||||||
| Projections & Slices |
| ||||||||||||
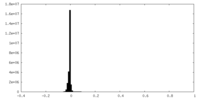
| Density Histograms |
-Half map: half map 1 of Nucleosome core particle from Trypanosoma brucei
| File | emd_16777_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 of Nucleosome core particle from Trypanosoma brucei | ||||||||||||
| Projections & Slices |
| ||||||||||||
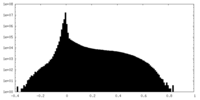
| Density Histograms |
- Sample components
Sample components
-Entire : Nucleosome core particle
| Entire | Name: Nucleosome core particle |
|---|---|
| Components |
|
-Supramolecule #1: Nucleosome core particle
| Supramolecule | Name: Nucleosome core particle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Nucleosome core particle with histones from Trypanosoma brucei and Widom 601 145 bp DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 89.6 KDa |
-Supramolecule #2: Histone octamer
| Supramolecule | Name: Histone octamer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 Details: Trypanosoma brucei histone octamer composed of an H4-H3-H3-H4 tetramer and two H2A-H2B dimers |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Widom 601 145 bp DNA
| Supramolecule | Name: Widom 601 145 bp DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5-#6 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Histone H3, putative
| Macromolecule | Name: Histone H3, putative / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.647858 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SRTKETARTK KTITSKKSKK ASKGSDAASG VKTAQRRWRP GTVALREIRQ FQRSTDLLLQ KAPFQRLVRE VSGAQKEGLR FQSSAILAA QEATESYIVS LLADTNRACI HSGRVTIQPK DIHLALCLRG ERA UniProtKB: Histone H3 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.037914 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AKGKKSGEAK GSQKRQKKVL RENVRGITRG SIRRLARRGG VKRISGVIYD EVRGVLKSFV EGVVRDATAY TEYSRKKTVT AVDVVNALR KRGKILYGYA UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.108614 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ATPKQAVKKA SKGGSSRSVK AGLIFPVGRV GTLLRRGQYA RRIGASGAVY MAAVLEYLTA ELLELSVKAA AQQTKKTKRL TPRTVTLAV RHDDDLGALL RNVTMSRGGV MPSLNKALAK KQKSGKHAKA TPSV UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.464503 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ATPKSTPAKT RKEAKKTRRQ RKRTWNVYVS RSLRSINSQM SMTSRTMKIV NSFVNDLFER IAAEAATIVR VNRKRTLGAR ELQTAVRLV LPADLAKHAM AEGTKAVSHA SS UniProtKB: Histone H2B |
-Macromolecule #5: Widom 601 145 bp DNA (127-mer ordered and built)
| Macromolecule | Name: Widom 601 145 bp DNA (127-mer ordered and built) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 44.99166 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC)(DC) ...String: (DA)(DT)(DC)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC)(DC)(DC)(DC) (DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT)(DA) (DA)(DA) (DA)(DC)(DG)(DC)(DG)(DG)(DG) (DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC)(DG) (DT)(DA)(DC) (DG)(DT)(DG)(DC)(DG)(DT) (DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG) (DA)(DG)(DC)(DT)(DG) (DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC)(DA) (DA)(DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG) (DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC)(DC) (DG)(DG)(DG)(DA)(DT)(DT) (DC)(DT)(DG) (DA)(DT) |
-Macromolecule #6: Widom 601 145 bp DNA (127-mer ordered and built)
| Macromolecule | Name: Widom 601 145 bp DNA (127-mer ordered and built) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 44.520383 KDa |
| Sequence | String: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC) ...String: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA) (DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC) (DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG) (DG)(DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT)(DC) (DT)(DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT)(DA) (DT)(DA)(DT)(DA)(DC)(DA) (DT)(DC)(DG) (DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 3e-05 kPa / Details: PELCO easiglow |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Glutaraldehyde fixation, S200 purification |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4913 / Average exposure time: 7.0 sec. / Average electron dose: 45.7 e/Å2 Details: correlated double sampling used, super-resolution, binning at motion correction stage |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 75.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)