+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Type II Secretion System | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type II Secretion System / MEMBRANE PROTEIN / Cell envelope | |||||||||
| Function / homology |  Function and homology information Function and homology informationtype II protein secretion system complex / cell envelope / protein secretion Similarity search - Function | |||||||||
| Biological species |  Deinococcus radiodurans R1 (radioresistant) / Deinococcus radiodurans R1 (radioresistant) /  Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) | |||||||||
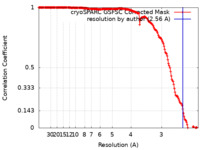
| Method | single particle reconstruction / cryo EM / Resolution: 2.56 Å | |||||||||
 Authors Authors | Farci D / Piano D | |||||||||
| Funding support |  Poland, 2 items Poland, 2 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: Structural characterization and functional insights into the type II secretion system of the poly-extremophile Deinococcus radiodurans. Authors: Domenica Farci / Stefan Milenkovic / Luca Iesu / Marta Tanas / Matteo Ceccarelli / Dario Piano /  Abstract: The extremophile bacterium D. radiodurans boasts a distinctive cell envelope characterized by the regular arrangement of three protein complexes. Among these, the Type II Secretion System (T2SS) ...The extremophile bacterium D. radiodurans boasts a distinctive cell envelope characterized by the regular arrangement of three protein complexes. Among these, the Type II Secretion System (T2SS) stands out as a pivotal structural component. We used cryo-electron microscopy to reveal unique features, such as an unconventional protein belt (DR_1364) around the main secretin (GspD), and a cap (DR_0940) found to be a separated subunit rather than integrated with GspD. Furthermore, a novel region at the N-terminus of the GspD constitutes an additional second gate, supplementing the one typically found in the outer membrane region. This T2SS was found to contribute to envelope integrity, while also playing a role in nucleic acid and nutrient trafficking. Studies on intact cell envelopes show a consistent T2SS structure repetition, highlighting its significance within the cellular framework. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16770.map.gz emd_16770.map.gz | 17.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16770-v30.xml emd-16770-v30.xml emd-16770.xml emd-16770.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16770_fsc.xml emd_16770_fsc.xml | 18.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_16770.png emd_16770.png | 85.2 KB | ||
| Filedesc metadata |  emd-16770.cif.gz emd-16770.cif.gz | 6.1 KB | ||
| Others |  emd_16770_half_map_1.map.gz emd_16770_half_map_1.map.gz emd_16770_half_map_2.map.gz emd_16770_half_map_2.map.gz | 442 MB 442 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16770 http://ftp.pdbj.org/pub/emdb/structures/EMD-16770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16770 | HTTPS FTP |
-Related structure data
| Related structure data |  8co1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16770.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16770.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
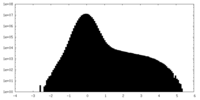
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||
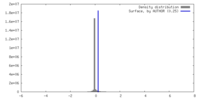
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B; same hand as primary map
| File | emd_16770_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B; same hand as primary map | ||||||||||||
| Projections & Slices |
| ||||||||||||
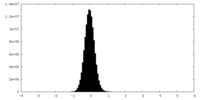
| Density Histograms |
-Half map: half map A; same hand as primary map
| File | emd_16770_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A; same hand as primary map | ||||||||||||
| Projections & Slices |
| ||||||||||||
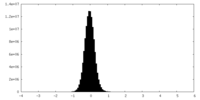
| Density Histograms |
- Sample components
Sample components
-Entire : Type II Secretion System
| Entire | Name: Type II Secretion System |
|---|---|
| Components |
|
-Supramolecule #1: Type II Secretion System
| Supramolecule | Name: Type II Secretion System / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 (radioresistant) Deinococcus radiodurans R1 (radioresistant) |
| Molecular weight | Theoretical: 1.7 MDa |
-Macromolecule #1: Probable type IV piliation system protein DR_0774
| Macromolecule | Name: Probable type IV piliation system protein DR_0774 / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) |
| Molecular weight | Theoretical: 78.653461 KDa |
| Sequence | String: MNKRHALLLT AVLGMATAYA QTAPTTTTVN TLQTVYRDPS LTSAPITANV GKYVGPLSTF LASIAKSAGY EVVFNFNIDA LALINGEIV FGNSTASVTT SYATPLGRPQ ELPAKPVVHN FSNAPFNEAW PLLMDVYELD YQLVKVGSAN VIRIGQRPKQ L ALPLKFIS ...String: MNKRHALLLT AVLGMATAYA QTAPTTTTVN TLQTVYRDPS LTSAPITANV GKYVGPLSTF LASIAKSAGY EVVFNFNIDA LALINGEIV FGNSTASVTT SYATPLGRPQ ELPAKPVVHN FSNAPFNEAW PLLMDVYELD YQLVKVGSAN VIRIGQRPKQ L ALPLKFIS AESALTAIEK FFGEEKFETV ISLDSNNKPF QTTRPTGKFG LPNSIKVIPD SSNKRLIIGS NSEDGIRIRS FV ETIDVQS SGKVISTDSI SEIYIVRGQK ESVLQFLRDS FPELIVTDYA SGGLAIEGPR TSVNRAIILL GQVDRAPEIP IVQ RIYTVR GQAADITALL AAQYPTLRVT PVGQTGQLVL NGAQAQLDTA LALLEQVDRP APVAESRTVQ RVFQLVNASA EEVK ATLEG TLARDLTADS NNDVLPNVPV TATDANGNTT VVSVPNALGK TANQGTANAQ AQTAQTPANT QQATLIADKR TNSLI VRGT PEQVAQVAEL VPQLDQVVPQ INVQVRIQEV NERALQSLGL NWRATFGGFN VAVSGGTGLA ATFNPTQSFL GFNIFP TLT ALETQGLTRR VYDGNVTMQS GQRSLSATGG AQNASSGAAA SVKSGGRLEI NIPSAAGNIV RQIDYGLNLD FFSPQVA PD GTITLRIRGQ VNQPATAITA DSLPNLIDFT NSEAQSTITF KNGQTILMSG LLGSTETTNR SGVPFLSSLP GVGAAFGE K RTEKTQSQLL VIITGTVVK UniProtKB: Probable type IV piliation system protein DR_0774 |
-Macromolecule #2: IPT/TIG domain-containing protein
| Macromolecule | Name: IPT/TIG domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) |
| Molecular weight | Theoretical: 15.954317 KDa |
| Sequence | String: MTALGVEHDQ HPACGGGSLT RHLQLIRLPG GGLSMLRFFC ASLLLTGLLA SCTPRVTTVA GVTVTPVLIK VSEGAAPGDT LTIQGRYLG NAQTARVIIG ADENGQGGTA FPASAVQSWS DTEIVLKVPE GMPAGGSWLF VEVGGKRSTG LRVSVR UniProtKB: IPT/TIG domain-containing protein |
-Macromolecule #3: Lipoprotein
| Macromolecule | Name: Lipoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 (radioresistant) |
| Molecular weight | Theoretical: 21.814654 KDa |
| Sequence | String: MTMKKMFPVL LLGGLLLAGC GTVGLGSGRV NVGVDVGDAG SEQVATLTIT PEKCDDKGVC TPQKQELSIT DGQPVTFTFT ARPGSEAVT IEGYRVLSDR LDGVERADPK NPVENAKMNL YVPSGYACEG LTAGASCQGN ESDIRIANGQ PVQHQIYFAS G LGARAAAK ...String: MTMKKMFPVL LLGGLLLAGC GTVGLGSGRV NVGVDVGDAG SEQVATLTIT PEKCDDKGVC TPQKQELSIT DGQPVTFTFT ARPGSEAVT IEGYRVLSDR LDGVERADPK NPVENAKMNL YVPSGYACEG LTAGASCQGN ESDIRIANGQ PVQHQIYFAS G LGARAAAK GANVTRVVDL EFYGFSANNV PFTRKVTGIV SQGSYVVKTN UniProtKB: Lipoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)