+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
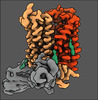
| Title | PHT1 in the outward facing conformation, bound to Sb27 | |||||||||
 Map data Map data | Map sharpened in cryoSPARC. Used for final model refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PHT1 / Peptide transporter / Histidine transporter / Sybody 27 / SLE / SLC15A4 / MEMBRANE PROTEIN | |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
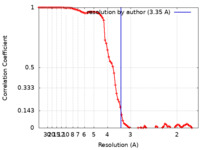
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Custodio T / Killer M / Loew C | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Molecular basis of TASL recruitment by the peptide/histidine transporter 1, PHT1. Authors: Tânia F Custódio / Maxime Killer / Dingquan Yu / Virginia Puente / Daniel P Teufel / Alexander Pautsch / Gisela Schnapp / Marc Grundl / Jan Kosinski / Christian Löw /  Abstract: PHT1 is a histidine /oligopeptide transporter with an essential role in Toll-like receptor innate immune responses. It can act as a receptor by recruiting the adaptor protein TASL which leads to type ...PHT1 is a histidine /oligopeptide transporter with an essential role in Toll-like receptor innate immune responses. It can act as a receptor by recruiting the adaptor protein TASL which leads to type I interferon production via IRF5. Persistent stimulation of this signalling pathway is known to be involved in the pathogenesis of systemic lupus erythematosus (SLE). Understanding how PHT1 recruits TASL at the molecular level, is therefore clinically important for the development of therapeutics against SLE and other autoimmune diseases. Here we present the Cryo-EM structure of PHT1 stabilized in the outward-open conformation. By combining biochemical and structural modeling techniques we propose a model of the PHT1-TASL complex, in which the first 16 N-terminal TASL residues fold into a helical structure that bind in the central cavity of the inward-open conformation of PHT1. This work provides critical insights into the molecular basis of PHT1/TASL mediated type I interferon production. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16758.map.gz emd_16758.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16758-v30.xml emd-16758-v30.xml emd-16758.xml emd-16758.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16758_fsc.xml emd_16758_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_16758.png emd_16758.png | 149 KB | ||
| Masks |  emd_16758_msk_1.map emd_16758_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16758.cif.gz emd-16758.cif.gz | 6 KB | ||
| Others |  emd_16758_additional_1.map.gz emd_16758_additional_1.map.gz emd_16758_additional_2.map.gz emd_16758_additional_2.map.gz emd_16758_half_map_1.map.gz emd_16758_half_map_1.map.gz emd_16758_half_map_2.map.gz emd_16758_half_map_2.map.gz | 56.8 MB 30.9 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16758 http://ftp.pdbj.org/pub/emdb/structures/EMD-16758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16758 | HTTPS FTP |
-Validation report
| Summary document |  emd_16758_validation.pdf.gz emd_16758_validation.pdf.gz | 789.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16758_full_validation.pdf.gz emd_16758_full_validation.pdf.gz | 789.3 KB | Display | |
| Data in XML |  emd_16758_validation.xml.gz emd_16758_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_16758_validation.cif.gz emd_16758_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16758 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16758 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16758 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16758 | HTTPS FTP |
-Related structure data
| Related structure data |  8cniMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16758.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16758.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened in cryoSPARC. Used for final model refinement. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
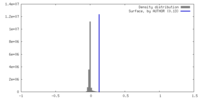
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16758_msk_1.map emd_16758_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
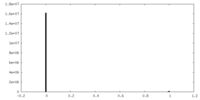
| Density Histograms |
-Additional map: Map post processed with DeepEMhancer. Tight Target model....
| File | emd_16758_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map post processed with DeepEMhancer. Tight Target model. Used for display and initial docking of sybody 27 only. | ||||||||||||
| Projections & Slices |
| ||||||||||||
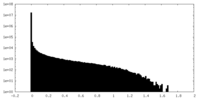
| Density Histograms |
-Additional map: Unsharpened full map. Final Non Uniform Refinement in cryoSPARC.
| File | emd_16758_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened full map. Final Non Uniform Refinement in cryoSPARC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
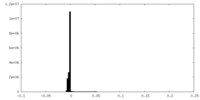
| Density Histograms |
-Half map: half map A
| File | emd_16758_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_16758_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PHT1-Sb27
| Entire | Name: PHT1-Sb27 |
|---|---|
| Components |
|
-Supramolecule #1: PHT1-Sb27
| Supramolecule | Name: PHT1-Sb27 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Solute carrier family 15 member 4
| Supramolecule | Name: Solute carrier family 15 member 4 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Sybody 27
| Supramolecule | Name: Sybody 27 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Solute carrier family 15 member 4
| Macromolecule | Name: Solute carrier family 15 member 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.566996 KDa |
| Recombinant expression | Organism:  Homo sapiens subsp. 'Denisova' (Denisova hominin) Homo sapiens subsp. 'Denisova' (Denisova hominin) |
| Sequence | String: MEEAEQSERS PLLGSGERGR AAIGAFHGRR LACAAVLLAE LLERVAFYGI TSNLVLFLNG PPYDWEGAQA SQALLLFMGI TYLVSPFGG WLADALLGKF GTILLSMALY LLGMLAFPVI AAPHTRQGLC GDIPLYPVEN CSSPATNATL APCSQVGTTR Y CAAATFVG ...String: MEEAEQSERS PLLGSGERGR AAIGAFHGRR LACAAVLLAE LLERVAFYGI TSNLVLFLNG PPYDWEGAQA SQALLLFMGI TYLVSPFGG WLADALLGKF GTILLSMALY LLGMLAFPVI AAPHTRQGLC GDIPLYPVEN CSSPATNATL APCSQVGTTR Y CAAATFVG LVLVGLGVGS VKANITPFGA DQVKDRGPEA TRRFFNWFYW SINLGAILSL GGIAYIQQNV SFVIGYSIPA IC IGISFMV FLCGQSFFIT KPPDGSAFTD MFKILAYSCC SRKRHMEHST NSEGQGVLQQ PRKQSLFEMA KLSRGGPFRE DKV EDVKAL VKIIPVFLAL IPYWTVYFQM QTTYVLQSLH LKIPEIANDT NSVHTFPAAW LTMFDAVLIL ILIPLKDKLV DPVL KRNGL LPSSLKRIAV GMFFVMCSAF AAGILESNRL KIVKVKTINQ TIGNVTYHAA DMPIWWQIPQ YVLIGFSEIF ASIAG LEFA YSAAPKSMQS AIMGLFFFFS GIGSFVGSGL LALVSIKEIG WMSNHTDFGN INGCQLNYYF FLLAAIQGAT LLLFLI VSV KYDHQKSKMN DVAANGRI UniProtKB: UNIPROTKB: F1NG54 |
-Macromolecule #2: Sybody 27
| Macromolecule | Name: Sybody 27 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.504001 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG SVQAGGSLRL SCAASGDIET IWYLGWFRQA PGKEREGVAA LSTVTGSTYY ADSVKGRFTV SLDNAKNTVY LQMNSLKPE DTALYYCAAA YTGWMAPLWQ WVYSYWGQGT QVTVS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: experimental model |
|---|---|
| Output model |  PDB-8cni: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)