[English] 日本語
 Yorodumi
Yorodumi- EMDB-16656: Low-resolution structure of the NorQ chaperone from Paracoccus de... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Low-resolution structure of the NorQ chaperone from Paracoccus denitrificans | ||||||||||||
 Map data Map data | Low-resolution structure of the NorQ chaperone from Paracoccus denitrificans | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | MoxR AAA+ and VWA domain proteins / CHAPERONE | ||||||||||||
| Biological species |  Paracoccus denitrificans (bacteria) Paracoccus denitrificans (bacteria) | ||||||||||||
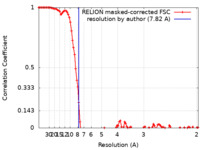
| Method | single particle reconstruction / cryo EM / Resolution: 7.82 Å | ||||||||||||
 Authors Authors | Adelroth P / Carroni M / Kahle M / Appelgren S / Elofsson A | ||||||||||||
| Funding support |  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: BMC Biol / Year: 2023 Journal: BMC Biol / Year: 2023Title: Insights into the structure-function relationship of the NorQ/NorD chaperones from Paracoccus denitrificans reveal shared principles of interacting MoxR AAA+/VWA domain proteins. Authors: Maximilian Kahle / Sofia Appelgren / Arne Elofsson / Marta Carroni / Pia Ädelroth /   Abstract: BACKGROUND: NorQ, a member of the MoxR-class of AAA+ ATPases, and NorD, a protein containing a Von Willebrand Factor Type A (VWA) domain, are essential for non-heme iron (Fe) cofactor insertion into ...BACKGROUND: NorQ, a member of the MoxR-class of AAA+ ATPases, and NorD, a protein containing a Von Willebrand Factor Type A (VWA) domain, are essential for non-heme iron (Fe) cofactor insertion into cytochrome c-dependent nitric oxide reductase (cNOR). cNOR catalyzes NO reduction, a key step of bacterial denitrification. This work aimed at elucidating the specific mechanism of NorQD-catalyzed Fe insertion, and the general mechanism of the MoxR/VWA interacting protein families. RESULTS: We show that NorQ-catalyzed ATP hydrolysis, an intact VWA domain in NorD, and specific surface carboxylates on cNOR are all features required for cNOR activation. Supported by BN-PAGE, low- ...RESULTS: We show that NorQ-catalyzed ATP hydrolysis, an intact VWA domain in NorD, and specific surface carboxylates on cNOR are all features required for cNOR activation. Supported by BN-PAGE, low-resolution cryo-EM structures of NorQ and the NorQD complex show that NorQ forms a circular hexamer with a monomer of NorD binding both to the side and to the central pore of the NorQ ring. Guided by AlphaFold predictions, we assign the density that "plugs" the NorQ ring pore to the VWA domain of NorD with a protruding "finger" inserting through the pore and suggest this binding mode to be general for MoxR/VWA couples. CONCLUSIONS: Based on our results, we present a tentative model for the mechanism of NorQD-catalyzed cNOR remodeling and suggest many of its features to be applicable to the whole MoxR/VWA family. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16656.map.gz emd_16656.map.gz | 78.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16656-v30.xml emd-16656-v30.xml emd-16656.xml emd-16656.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16656_fsc.xml emd_16656_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16656.png emd_16656.png | 78 KB | ||
| Masks |  emd_16656_msk_1.map emd_16656_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_16656_half_map_1.map.gz emd_16656_half_map_1.map.gz emd_16656_half_map_2.map.gz emd_16656_half_map_2.map.gz | 79.4 MB 79.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16656 http://ftp.pdbj.org/pub/emdb/structures/EMD-16656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16656 | HTTPS FTP |
-Validation report
| Summary document |  emd_16656_validation.pdf.gz emd_16656_validation.pdf.gz | 665.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16656_full_validation.pdf.gz emd_16656_full_validation.pdf.gz | 665.5 KB | Display | |
| Data in XML |  emd_16656_validation.xml.gz emd_16656_validation.xml.gz | 17.8 KB | Display | |
| Data in CIF |  emd_16656_validation.cif.gz emd_16656_validation.cif.gz | 23.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16656 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16656 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16656 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16656 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16656.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16656.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Low-resolution structure of the NorQ chaperone from Paracoccus denitrificans | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.99 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16656_msk_1.map emd_16656_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
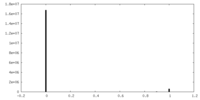
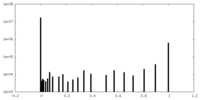
| Density Histograms |
-Half map: Low-resolution structure of the NorQ chaperone from Paracoccus...
| File | emd_16656_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Low-resolution structure of the NorQ chaperone from Paracoccus denitrificans Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
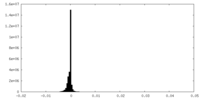
| Density Histograms |
-Half map: Low-resolution structure of the NorQ chaperone from Paracoccus...
| File | emd_16656_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Low-resolution structure of the NorQ chaperone from Paracoccus denitrificans Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
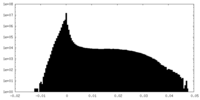
| Density Histograms |
- Sample components
Sample components
-Entire : NorQ hexamer
| Entire | Name: NorQ hexamer |
|---|---|
| Components |
|
-Supramolecule #1: NorQ hexamer
| Supramolecule | Name: NorQ hexamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Paracoccus denitrificans (bacteria) Paracoccus denitrificans (bacteria) |
-Macromolecule #1: Pd. NorQ
| Macromolecule | Name: Pd. NorQ / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Paracoccus denitrificans (bacteria) Paracoccus denitrificans (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHASN AHVKTQGNGA VDAPFYLPQG DEVAVFEAAA ANDLPVLLKG PTGCGKTRFV AHMAARLG R PLYTVACHDD LSAADLIGRY LLKGGETVWT DGPLTRAVRE GAICYLDQVV EARKDVTVV LHPLTDDRRI LPIDRTGEEI EAAPGFMLVA SYNPGYQNIL ...String: MHHHHHHASN AHVKTQGNGA VDAPFYLPQG DEVAVFEAAA ANDLPVLLKG PTGCGKTRFV AHMAARLG R PLYTVACHDD LSAADLIGRY LLKGGETVWT DGPLTRAVRE GAICYLDQVV EARKDVTVV LHPLTDDRRI LPIDRTGEEI EAAPGFMLVA SYNPGYQNIL KTLKPSTRQR FVAMEFDFPE PAREVEIVA RESGLDRDRT LGLVRLAGKI RGLKGQDLEE GVSTRLVVYA ASLTRRGMNL D RAIEAAMI EPLTDDAEVK RGLRDLAAAI FG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 20 mM HEPES, 150 mM NaCl, 2 mM DTT |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)