[English] 日本語
 Yorodumi
Yorodumi- EMDB-1664: Alpha-helical nascent polypeptide chains visualized within distin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1664 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
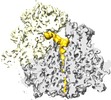
| Title | Alpha-helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel | |||||||||
 Map data Map data | 80S.Helix1_RNC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / Protein exit tunnel / co-translational protein folding | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 7.1 Å | |||||||||
 Authors Authors | Bhushan S / Gartmann M / Halic M / Armache JP / Jarasch A / Mielke T / Berninghausen O / Wilson DN / Beckmann R | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2010 Journal: Nat Struct Mol Biol / Year: 2010Title: alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Authors: Shashi Bhushan / Marco Gartmann / Mario Halic / Jean-Paul Armache / Alexander Jarasch / Thorsten Mielke / Otto Berninghausen / Daniel N Wilson / Roland Beckmann /  Abstract: As translation proceeds, the nascent polypeptide chain passes through a tunnel in the large ribosomal subunit. Although this ribosomal exit tunnel was once thought only to be a passive conduit for ...As translation proceeds, the nascent polypeptide chain passes through a tunnel in the large ribosomal subunit. Although this ribosomal exit tunnel was once thought only to be a passive conduit for the growing nascent chain, accumulating evidence suggests that it may in fact play a more active role in regulating translation and initial protein folding events. Here we have determined single-particle cryo-electron microscopy reconstructions of eukaryotic 80S ribosomes containing nascent chains with high alpha-helical propensity located within the exit tunnel. The maps enable direct visualization of density for helices as well as allowing the sites of interaction with the tunnel wall components to be elucidated. In particular regions of the tunnel, the nascent chain adopts distinct conformations and establishes specific contacts with tunnel components, both ribosomal RNA and proteins, that have been previously implicated in nascent chain-ribosome interaction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1664.map.gz emd_1664.map.gz | 7.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1664-v30.xml emd-1664-v30.xml emd-1664.xml emd-1664.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1664.png EMD-1664.png | 205.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1664 http://ftp.pdbj.org/pub/emdb/structures/EMD-1664 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1664 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1664 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1664.map.gz / Format: CCP4 / Size: 185.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1664.map.gz / Format: CCP4 / Size: 185.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 80S.Helix1_RNC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
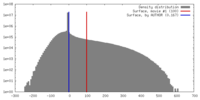
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
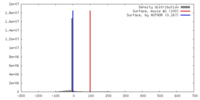
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : This map represents a wheat germ 80S ribosomal nascent chain comp...
| Entire | Name: This map represents a wheat germ 80S ribosomal nascent chain complex with different nascent chains |
|---|---|
| Components |
|
-Supramolecule #1000: This map represents a wheat germ 80S ribosomal nascent chain comp...
| Supramolecule | Name: This map represents a wheat germ 80S ribosomal nascent chain complex with different nascent chains type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 4.1 MDa / Theoretical: 4.1 MDa / Method: Known for 80S ribosomes |
-Supramolecule #1: Ribosome
| Supramolecule | Name: Ribosome / type: complex / ID: 1 / Name.synonym: Ribosome / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 4.1 MDa / Theoretical: 4.1 MDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Hepes/KOH ph 7.5, 100 mM KOAc, 10 mM Mg(oAc)2, 1.5 mM DTT, 0.1% (w/v) Nikol |
| Staining | Type: NEGATIVE / Details: Cryo-EM |
| Grid | Details: Quantifoil grids (3/3) with 2nm carbon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: OTHER / Details: Vitrification instrument: Vitrobot / Method: Blot for 10 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Average: 84 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 4.76 µm / Number real images: 150 / Average electron dose: 25 e/Å2 / Details: Scanned at 5334 dpi / Od range: 1.2 / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 38000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 39000 |
| Sample stage | Specimen holder: FEI Polara cartridge system / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Signature for particle selection and web for visual inspection |
|---|---|
| CTF correction | Details: Defocus group volumes |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.1 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 120000 |
| Final angle assignment | Details: SPIDER |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)