[English] 日本語
 Yorodumi
Yorodumi- EMDB-16631: The ACP crosslinked to the KS of the cercosporin fungal non-reduc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The ACP crosslinked to the KS of the cercosporin fungal non-reducing polyketide synthase (NR-PKS) CTB1 (SAT-KS:ACP-MAT) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fungal non-reducing polyketide synthase (NR-PKS) / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsecondary metabolite biosynthetic process / fatty acid synthase activity / phosphopantetheine binding / 3-oxoacyl-[acyl-carrier-protein] synthase activity / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / fatty acid biosynthetic process Similarity search - Function | |||||||||
| Biological species |  Cercospora nicotianae (fungus) Cercospora nicotianae (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Munoz-Hernandez H / Tittes YU / Maier T | |||||||||
| Funding support | European Union,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of ACP crosslinked to KS of the cercosporin fungal non-reducing polyketide synthase (NR-PKS) CTB1 (SAT-KS:ACP-MAT) at 2.7 Angstroms resolution Authors: Munoz-Hernandez H / Maier T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16631.map.gz emd_16631.map.gz | 108.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16631-v30.xml emd-16631-v30.xml emd-16631.xml emd-16631.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16631.png emd_16631.png | 60.5 KB | ||
| Filedesc metadata |  emd-16631.cif.gz emd-16631.cif.gz | 6.6 KB | ||
| Others |  emd_16631_half_map_1.map.gz emd_16631_half_map_1.map.gz emd_16631_half_map_2.map.gz emd_16631_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16631 http://ftp.pdbj.org/pub/emdb/structures/EMD-16631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16631 | HTTPS FTP |
-Validation report
| Summary document |  emd_16631_validation.pdf.gz emd_16631_validation.pdf.gz | 981.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16631_full_validation.pdf.gz emd_16631_full_validation.pdf.gz | 981.1 KB | Display | |
| Data in XML |  emd_16631_validation.xml.gz emd_16631_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_16631_validation.cif.gz emd_16631_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16631 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16631 | HTTPS FTP |
-Related structure data
| Related structure data |  8cg5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16631.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16631.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
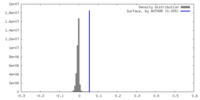
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_16631_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
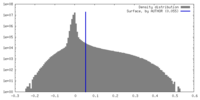
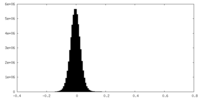
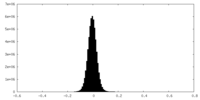
| Density Histograms |
-Half map: #1
| File | emd_16631_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
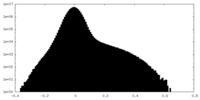
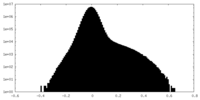
| Density Histograms |
- Sample components
Sample components
-Entire : ACP crosslinked to the KS of the condensing region of the non-red...
| Entire | Name: ACP crosslinked to the KS of the condensing region of the non-reducing polyketide synthase CTB1 |
|---|---|
| Components |
|
-Supramolecule #1: ACP crosslinked to the KS of the condensing region of the non-red...
| Supramolecule | Name: ACP crosslinked to the KS of the condensing region of the non-reducing polyketide synthase CTB1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Cercospora nicotianae (fungus) Cercospora nicotianae (fungus) |
-Macromolecule #1: Non-reducing polyketide synthase CTB1
| Macromolecule | Name: Non-reducing polyketide synthase CTB1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  Cercospora nicotianae (fungus) Cercospora nicotianae (fungus) |
| Molecular weight | Theoretical: 140.255 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEDGAQMRVV AFGDQTYDCS EAVSQLLRVR DDAIVVDFLE RAPAVLKAEL ARLSSEQQEE TPRFATLAEL VPRYRAGTLN PAVSQALTC IAQLGLFIRQ HSSGQEAYPT AHDSCITGVA TGALTAVAVG SASSVTALVP LALHTVAVAV RLGARAWEIG S CLADARRG ...String: MEDGAQMRVV AFGDQTYDCS EAVSQLLRVR DDAIVVDFLE RAPAVLKAEL ARLSSEQQEE TPRFATLAEL VPRYRAGTLN PAVSQALTC IAQLGLFIRQ HSSGQEAYPT AHDSCITGVA TGALTAVAVG SASSVTALVP LALHTVAVAV RLGARAWEIG S CLADARRG ANGRYASWTS AVGGISPQDL QDRISAYTAE QALASVSVPY LSAAVGPGQS SVSAAPVILD AFLSTLLRPL TT TRLPITA PYHAPHLFTA KDVQHVTDCL PPSEAWPTVR IPIISFSRDE AVSRGASFPA AMSEAVRDCL IRPIALDRMA VSI ANHARD LGKDSVLPSP IALSFSDKLG PQVNSHLPGA KAPTPELTSK SIPSAIGAEQ QPMAKSPIAI LAASGRFPQS SSMD QFWDV LINGVDTHEL VPPTRWNAAT HVSEDPKAKN VSGTGFGCWL HEAGEFDAAY FNMSPREAPQ VDPAQRLALL TATEA LEQA GVVPNRTSST QKNRVGVWYG ATSNDWMETN SAQNVDTYFI PGGNRAFIPG RVNYFHKFSG PSYTIDTACS SSLAAL HMA CNALWRGEVD TAIVGGTNVL TNPDMTAGLD AGHFLSRSGN CKTFDDEADG YCRGEAVVTL ILKRLPDAQA DKDPIQA SI LGIATNHSAE AASITRPHAG AQQDLFQQVL TETGLTANDI SVCEMHGTGT QAGDSGETTS VVETLAPLNR SGSAVRTT P LYIGAVKSNV GHAESAAGVS SLAKILLMLK HSKIPPHVGI KTKLNHRLPD LAARNTHIAR SEVPWPRPKN GKRRVLLNN FSAAGGNTCL VLEDAPEPED SQEVDPREHH IVALSAKTPD SMVNNLTNMI TWIDKHSGDS LATLPQLSYT TTARRVHHRH RAVATGTDL LQIRSSLQEQ LDRRVSGERS IPHPPNGPSF VLAFTGQGSA FAGMGVDLYK RFASFRSDIA RYDQICEGMS L PSIKAMFE DEKVFSTASP TLQQLTHVCF QMALYRLWKS LGVQAKAVVG HALGEYAALY AAGVLSQSDT LYLVGRRAQL ME KHLSQGT HAMLAVRAKE EAIVAAIDGP PGEAYEFSCR NGEQRNVLGG TVAQIQAAKA ALEAKKIRCQ YLDTPMAFHT GQV DPILPE LLQVAAACSI QDPQIPVISP AYGKVIRSAK DFQPEYFTHH CRSSVNMVDA LQSAVEEGLL DKNVIGLEIG PGPV VTQFV KEAVGTTMQT FASINKDKDT WQLMTQALAK FYLAGASVEW SRYHEDFPGA QKVLELPAYG WALKNYWLQY VNDWS LRKG DPAVVVAASA AALEHHHHHH UniProtKB: Non-reducing polyketide synthase CTB1 |
-Macromolecule #2: Acyl carrier protein (ACP) of Non-reducing polyketide synthase CTB1
| Macromolecule | Name: Acyl carrier protein (ACP) of Non-reducing polyketide synthase CTB1 type: protein_or_peptide / ID: 2 Details: Acyl carrier protein (ACP) of Non-reducing polyketide synthase CTB1 Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  Cercospora nicotianae (fungus) Cercospora nicotianae (fungus) |
| Molecular weight | Theoretical: 9.892837 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMDPSPNE IGTVWRDALK ILSEESGLTD EELTDDTSFA DVGVDSLMSL VITSRLRDEL DIDFPDRALF EECQTIFDLR KRFSGSTE UniProtKB: Non-reducing polyketide synthase CTB1 |
-Macromolecule #3: N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-[2-...
| Macromolecule | Name: N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-[2-(propanoylamino)ethyl]-beta-alaninamide type: ligand / ID: 3 / Number of copies: 1 / Formula: 42X |
|---|---|
| Molecular weight | Theoretical: 397.361 Da |
| Chemical component information | 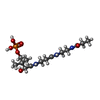 ChemComp-42X: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.3000000000000003 µm / Calibrated defocus min: 0.9 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8cg5: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)