[English] 日本語
 Yorodumi
Yorodumi- EMDB-16630: The organise full-length structure of the fungal non-reducing pol... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
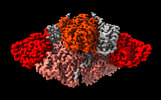
| Title | The organise full-length structure of the fungal non-reducing polyketide synthase (NR-PKS) PksA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fungal non-reducing polyketide synthase (NR-PKS) / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnoranthrone synthase / aflatoxin biosynthetic process / norsolorinate anthrone synthase activity / fatty acid synthase activity / phosphopantetheine binding / fatty acid biosynthetic process / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Munoz-Hernandez H / Maier T | |||||||||
| Funding support | European Union,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of the Aspergilus sp. fungal non-reducing polyketide synthase (NR-PKS) PksA at 2.6 Angstroms resolution Authors: Munoz-Hernandez H / Maier T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16630.map.gz emd_16630.map.gz | 107.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16630-v30.xml emd-16630-v30.xml emd-16630.xml emd-16630.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16630.png emd_16630.png | 78.2 KB | ||
| Filedesc metadata |  emd-16630.cif.gz emd-16630.cif.gz | 7.5 KB | ||
| Others |  emd_16630_half_map_1.map.gz emd_16630_half_map_1.map.gz emd_16630_half_map_2.map.gz emd_16630_half_map_2.map.gz | 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16630 http://ftp.pdbj.org/pub/emdb/structures/EMD-16630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16630 | HTTPS FTP |
-Validation report
| Summary document |  emd_16630_validation.pdf.gz emd_16630_validation.pdf.gz | 909.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16630_full_validation.pdf.gz emd_16630_full_validation.pdf.gz | 909 KB | Display | |
| Data in XML |  emd_16630_validation.xml.gz emd_16630_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_16630_validation.cif.gz emd_16630_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16630 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16630 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16630 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16630 | HTTPS FTP |
-Related structure data
| Related structure data |  8cg4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16630.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16630.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
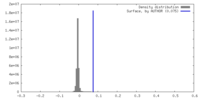
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16630_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
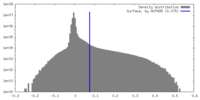
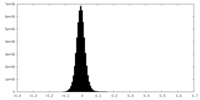
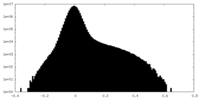
| Density Histograms |
-Half map: #2
| File | emd_16630_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
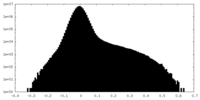
| Density Histograms |
- Sample components
Sample components
-Entire : The full-length Non-reducing polyketide synthase PksA
| Entire | Name: The full-length Non-reducing polyketide synthase PksA |
|---|---|
| Components |
|
-Supramolecule #1: The full-length Non-reducing polyketide synthase PksA
| Supramolecule | Name: The full-length Non-reducing polyketide synthase PksA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: Norsolorinic acid synthase
| Macromolecule | Name: Norsolorinic acid synthase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: noranthrone synthase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 237.74125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHHHHHHHH HHGSTSGSGE QKLISEEDLG STSGSGDYKD DDDKLTSLYK KAGLENLYFQ GMAQSRQLFL FGDQTADFVP KLRSLLSVQ DSPILAAFLD QSHYVVRAQM LQSMNTVDHK LARTADLRQM VQKYVDGKLT PAFRTALVCL CQLGCFIREY E ESGNMYPQ ...String: MAHHHHHHHH HHGSTSGSGE QKLISEEDLG STSGSGDYKD DDDKLTSLYK KAGLENLYFQ GMAQSRQLFL FGDQTADFVP KLRSLLSVQ DSPILAAFLD QSHYVVRAQM LQSMNTVDHK LARTADLRQM VQKYVDGKLT PAFRTALVCL CQLGCFIREY E ESGNMYPQ PSDSYVLGFC MGSLAAVAVS CSRSLSELLP IAVQTVLIAF RLGLCALEMR DRVDGCSDDR GDPWSTIVWG LD PQQARDQ IEVFCRTTNV PQTRRPWISC ISKNAITLSG SPSTLRAFCA MPQMAQHRTA PIPICLPAHN GALFTQADIT TIL DTTPTT PWEQLPGQIP YISHVTGNVV QTSNYRDLIE VALSETLLEQ VRLDLVETGL PRLLQSRQVK SVTIVPFLTR MNET MSNIL PDSFISTETR TDTGRAIPAS GRPGAGKCKL AIVSMSGRFP ESPTTESFWD LLYKGLDVCK EVPRRRWDIN THVDP SGKA RNKGATKWGC WLDFSGDFDP RFFGISPKEA PQMDPAQRMA LMSTYEAMER AGLVPDTTPS TQRDRIGVFH GVTSND WME TNTAQNIDTY FITGGNRGFI PGRINFCFEF AGPSYTNDTA CSSSLAAIHL ACNSLWRGDC DTAVAGGTNM IYTPDGH TG LDKGFFLSRT GNCKPYDDKA DGYCRAEGVG TVFIKRLEDA LADNDPILGV ILDAKTNHSA MSESMTRPHV GAQIDNMT A ALNTTGLHPN DFSYIEMHGT GTQVGDAVEM ESVLSVFAPS ETARKADQPL FVGSAKANVG HGEGVSGVTS LIKVLMMMQ HDTIPPHCGI KPGSKINRNF PDLGARNVHI AFEPKPWPRT HTPRRVLINN FSAAGGNTAL IVEDAPERHW PTEKDPRSSH IVALSAHVG ASMKTNLERL HQYLLKNPHT DLAQLSYTTT ARRWHYLHRV SVTGASVEEV TRKLEMAIQN GDGVSRPKSK P KILFAFTG QGSQYATMGK QVYDAYPSFR EDLEKFDRLA QSHGFPSFLH VCTSPKGDVE EMAPVVVQLA ITCLQMALTN LM TSFGIRP DVTVGHSLGE FAALYAAGVL SASDVVYLVG QRAELLQERC QRGTHAMLAV KATPEALSQW IQDHDCEVAC ING PEDTVL SGTTKNVAEV QRAMTDNGIK CTLLKLPFAF HSAQVQPILD DFEALAQGAT FAKPQLLILS PLLRTEIHEQ GVVT PSYVA QHCRHTVDMA QALRSAREKG LIDDKTLVIE LGPKPLISGM VKMTLGDKIS TLPTLAPNKA IWPSLQKILT SVYTG GWDI NWKKYHAPFA SSQKVVDLPS YGWDLKDYYI PYQGDWCLHR HQQDCKCAAP GHEIKTADYQ VPPESTPHRP SKLDPS KEA FPEIKTTTTL HRVVEETTKP LGATLVVETD ISRKDVNGLA RGHLVDGIPL CTPSFYADIA MQVGQYSMQR LRAGHPG AG AIDGLVDVSD MVVDKALVPH GKGPQLLRTT LTMEWPPKAA ATTRSAKVKF ATYFADGKLD TEHASCTVRF TSDAQLKS L RRSVSEYKTH IRQLHDGHAK GQFMRYNRKT GYKLMSSMAR FNPDYMLLDY LVLNEAENEA ASGVDFSLGS SEGTFAAHP AHVDAITQVA GFAMNANDNV DIEKQVYVNH GWDSFQIYQP LDNSKSYQVY TKMGQAKEND LVHGDVVVLD GEQIVAFFRG LTLRSVPRG ALRVVLQTTV KKADRQLGFK TMPSPPPPTT TMPISPYKPA NTQVSSQAIP AEATHSHTPP QPKHSPVPET A GSAPAAKG VGVSNEKLDA VMRVVSEESG IALEELTDDS NFADMGIDSL SSMVIGSRFR EDLGLDLGPE FSLFIDCTTV RA LKDFMLG SGDAGSGSNV EDPPPSATPG INPETDWSSS ASDSIFASED HGHSSESGAD TGSPPALDLK PYCRPSTSVV LQG LPMVAR KTLFMLPDGG GSAFSYASLP RLKSDTAVVG LNCPYARDPE NMNCTHGAMI ESFCNEIRRR QPRGPYHLGG WSSG GAFAY VVAEALVNQG EEVHSLIIID APIPQAMEQL PRAFYEHCNS IGLFATQPGA SPDGSTEPPS YLIPHFTAVV DVMLD YKLA PLHARRMPKV GIVWAADTVM DERDAPKMKG MHFMIQKRTE FGPDGWDTIM PGASFDIVRA DGANHFTLMQ KEHVSI ISD LIDRVMA UniProtKB: Norsolorinic acid synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Number grids imaged: 1 / Number real images: 6479 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.2 µm / Calibrated defocus min: 0.9 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8cg4: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)