[English] 日本語
 Yorodumi
Yorodumi- EMDB-16590: Structure of the yeast delta uL16m mitochondrial ribosome - State 2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the yeast delta uL16m mitochondrial ribosome - State 2 | |||||||||||||||
 Map data Map data | consensus map of delta uL16m yeast mitochondrial ribosome- State2 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Structure of the yeast delta uL16m mitochondrial ribosome / RIBOSOME | |||||||||||||||
| Biological species |  | |||||||||||||||
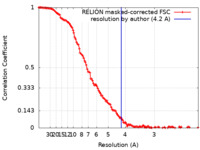
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||||||||
 Authors Authors | Conrad J / Rathore S / Barrientos A | |||||||||||||||
| Funding support |  United States, United States,  Sweden, Sweden,  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Biochim Biophys Acta Mol Cell Res / Year: 2025 Journal: Biochim Biophys Acta Mol Cell Res / Year: 2025Title: The late stages of yeast mitoribosome large subunit biogenesis. Authors: Sorbhi Rathore / Julian Conrad / Dasmanthie De Silva / Alberto Ferrari / Danielle Bouquio / Hyung-Jun Kim / Roger Salvatori / Andreas Linden / Olexandr Dybkov / Henning Urlaub / Martin Ott / ...Authors: Sorbhi Rathore / Julian Conrad / Dasmanthie De Silva / Alberto Ferrari / Danielle Bouquio / Hyung-Jun Kim / Roger Salvatori / Andreas Linden / Olexandr Dybkov / Henning Urlaub / Martin Ott / Antoni Barrientos /    Abstract: The Saccharomyces cerevisiae mitoribosome synthesizes eight mitochondrial DNA-encoded proteins essential for oxidative phosphorylation. Mitoribosome large subunit (mtLSU) biogenesis involves the ...The Saccharomyces cerevisiae mitoribosome synthesizes eight mitochondrial DNA-encoded proteins essential for oxidative phosphorylation. Mitoribosome large subunit (mtLSU) biogenesis involves the conserved DEAD-box helicase Mrh4 and the GTPases Mtg1/GTPBP7 and Mtg2/GTPBP5. Here, we have employed genetic, biochemical, in vitro reconstitution, and cryo-EM approaches to elucidate their hierarchical action during the late stages of mtLSU assembly. We show that Mrh4-mediated bL33m incorporation precedes Mtg1 recruitment to the 21S rRNA. Cryo-EM structures of mitoribosome assembly intermediates accumulating in the absence of Mtg1 or uL16m reveal that Mtg1 restructures the 21S rRNA H73-75 and H93 domains to their mature fold. This subsequently allows the structuring of neighboring peptidyl transfer center region helices and the incorporation of uL6m, uL16m, bL35m, and bL36m during late mtLSU maturation. Unexpectedly, monosomes containing immature mtLSU assemble in Mrh4-, bL33m-, uL16m-, Mtg1-, and Mtg2-depleted mitochondria, at levels that increase with the maturation state of the mtLSU particle. Our data have shed light on the rRNA folding events and the structuring of the MRPs that occur during the late stages of assembly. They have provided insight into the roles of assembly factors Mrh4, Mtg1, and Mtg2 during the process and revealed evolutionarily conserved mechanisms underlying mitochondrial ribosome assembly. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16590.map.gz emd_16590.map.gz | 160.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16590-v30.xml emd-16590-v30.xml emd-16590.xml emd-16590.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16590_fsc.xml emd_16590_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16590.png emd_16590.png | 152.2 KB | ||
| Filedesc metadata |  emd-16590.cif.gz emd-16590.cif.gz | 4.5 KB | ||
| Others |  emd_16590_additional_1.map.gz emd_16590_additional_1.map.gz emd_16590_additional_2.map.gz emd_16590_additional_2.map.gz emd_16590_additional_3.map.gz emd_16590_additional_3.map.gz emd_16590_half_map_1.map.gz emd_16590_half_map_1.map.gz emd_16590_half_map_2.map.gz emd_16590_half_map_2.map.gz | 46.4 MB 18.3 MB 47.8 MB 140.9 MB 140.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16590 http://ftp.pdbj.org/pub/emdb/structures/EMD-16590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16590 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16590.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16590.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | consensus map of delta uL16m yeast mitochondrial ribosome- State2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
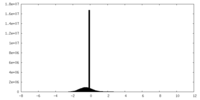
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Multibody refinement of delta uL16m yeast mitochondrial SSU body
| File | emd_16590_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Multibody refinement of delta uL16m yeast mitochondrial SSU body | ||||||||||||
| Projections & Slices |
| ||||||||||||
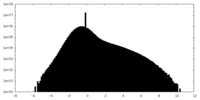
| Density Histograms |
-Additional map: Multibody refinement of delta uL16m yeast mitochondrial SSU head
| File | emd_16590_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Multibody refinement of delta uL16m yeast mitochondrial SSU head | ||||||||||||
| Projections & Slices |
| ||||||||||||
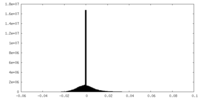
| Density Histograms |
-Additional map: Multibody refinement of delta uL16m yeast mitochondrial LSU
| File | emd_16590_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Multibody refinement of delta uL16m yeast mitochondrial LSU | ||||||||||||
| Projections & Slices |
| ||||||||||||
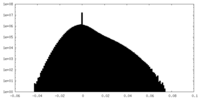
| Density Histograms |
-Half map: half map 2
| File | emd_16590_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_16590_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of the yeast delta uL16m mitochondrial ribosome - State 2
| Entire | Name: Structure of the yeast delta uL16m mitochondrial ribosome - State 2 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of the yeast delta uL16m mitochondrial ribosome - State 2
| Supramolecule | Name: Structure of the yeast delta uL16m mitochondrial ribosome - State 2 type: complex / ID: 1 / Parent: 0 Details: Mitoribosomal particles from an uL16m-deletion strain were purified by sucrose cushion sedimentation of mitochondrial extracts prepared in the presence of 20% Mg2+ and 1% Triton X-100. This ...Details: Mitoribosomal particles from an uL16m-deletion strain were purified by sucrose cushion sedimentation of mitochondrial extracts prepared in the presence of 20% Mg2+ and 1% Triton X-100. This approach revealed the structures of novel mitoribosome assembly intermediates. After processing, the structural characterization of the uL16m-deletion mitoribosome particles allowed us to define two major states: state 1, state 2. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)