+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tomogram of Serratia marcescens in the post-spanin state (2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Virulence / Secretion / Tc toxin / Type 10 secretion system / TOXIN | |||||||||
| Biological species |  Serratia marcescens (bacteria) Serratia marcescens (bacteria) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Sitsel O / Wang Z / Janning P / Kroczek L / Wagner T / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: Yersinia entomophaga Tc toxin is released by T10SS-dependent lysis of specialized cell subpopulations. Authors: Oleg Sitsel / Zhexin Wang / Petra Janning / Lara Kroczek / Thorsten Wagner / Stefan Raunser /  Abstract: Disease-causing bacteria secrete numerous toxins to invade and subjugate their hosts. Unlike many smaller toxins, the secretion machinery of most large toxins remains enigmatic. By combining genomic ...Disease-causing bacteria secrete numerous toxins to invade and subjugate their hosts. Unlike many smaller toxins, the secretion machinery of most large toxins remains enigmatic. By combining genomic editing, proteomic profiling and cryo-electron tomography of the insect pathogen Yersinia entomophaga, we demonstrate that a specialized subset of these cells produces a complex toxin cocktail, including the nearly ribosome-sized Tc toxin YenTc, which is subsequently exported by controlled cell lysis using a transcriptionally coupled, pH-dependent type 10 secretion system (T10SS). Our results dissect the Tc toxin export process by a T10SS, identifying that T10SSs operate via a previously unknown lytic mode of action and establishing them as crucial players in the size-insensitive release of cytoplasmically folded toxins. With T10SSs directly embedded in Tc toxin operons of major pathogens, we anticipate that our findings may model an important aspect of pathogenesis in bacteria with substantial impact on agriculture and healthcare. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16539.map.gz emd_16539.map.gz | 930.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16539-v30.xml emd-16539-v30.xml emd-16539.xml emd-16539.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16539.png emd_16539.png | 239.7 KB | ||
| Filedesc metadata |  emd-16539.cif.gz emd-16539.cif.gz | 4.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16539 http://ftp.pdbj.org/pub/emdb/structures/EMD-16539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16539 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16539.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16539.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 9.292 Å | ||||||||||||||||||||||||||||||||
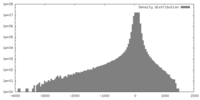
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Serratia marcescens strain BS 303, Ara-ChiR
| Entire | Name: Serratia marcescens strain BS 303, Ara-ChiR |
|---|---|
| Components |
|
-Supramolecule #1: Serratia marcescens strain BS 303, Ara-ChiR
| Supramolecule | Name: Serratia marcescens strain BS 303, Ara-ChiR / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Serratia marcescens (bacteria) / Strain: BS 303, Ara-ChiR Serratia marcescens (bacteria) / Strain: BS 303, Ara-ChiR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6 / Details: Directly frozen in SOC growth media |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: Aurion / Diameter: 5 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 114.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 41 |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)