[English] 日本語
 Yorodumi
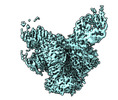
Yorodumi- EMDB-16493: cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | ||||||||||||
 Map data Map data | Overall cryo-EM map of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV / post-treatment controller / broadly neutralizing antibodies / V3-site / IMMUNE SYSTEM | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Baquero E / Molinos-Albert L / Mouquet H | ||||||||||||
| Funding support |  France, France,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2023 Journal: Cell Host Microbe / Year: 2023Title: Anti-V1/V3-glycan broadly HIV-1 neutralizing antibodies in a post-treatment controller. Authors: Luis M Molinos-Albert / Eduard Baquero / Mélanie Bouvin-Pley / Valérie Lorin / Caroline Charre / Cyril Planchais / Jordan D Dimitrov / Valérie Monceaux / Matthijn Vos / / Laurent ...Authors: Luis M Molinos-Albert / Eduard Baquero / Mélanie Bouvin-Pley / Valérie Lorin / Caroline Charre / Cyril Planchais / Jordan D Dimitrov / Valérie Monceaux / Matthijn Vos / / Laurent Hocqueloux / Jean-Luc Berger / Michael S Seaman / Martine Braibant / Véronique Avettand-Fenoël / Asier Sáez-Cirión / Hugo Mouquet /   Abstract: HIV-1 broadly neutralizing antibodies (bNAbs) can decrease viremia but are usually unable to counteract autologous viruses escaping the antibody pressure. Nonetheless, bNAbs may contribute to natural ...HIV-1 broadly neutralizing antibodies (bNAbs) can decrease viremia but are usually unable to counteract autologous viruses escaping the antibody pressure. Nonetheless, bNAbs may contribute to natural HIV-1 control in individuals off antiretroviral therapy (ART). Here, we describe a bNAb B cell lineage elicited in a post-treatment controller (PTC) that exhibits broad seroneutralization and show that a representative antibody from this lineage, EPTC112, targets a quaternary epitope in the glycan-V3 loop supersite of the HIV-1 envelope glycoprotein. The cryo-EM structure of EPTC112 complexed with soluble BG505 SOSIP.664 envelope trimers revealed interactions with N301- and N156-branched N-glycans and the GDIR V3 loop motif. Although the sole contemporaneous virus circulating in this PTC was resistant to EPTC112, it was potently neutralized by autologous plasma IgG antibodies. Our findings illuminate how cross-neutralizing antibodies can alter the HIV-1 infection course in PTCs and may control viremia off-ART, supporting their role in functional HIV-1 cure strategies. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16493.map.gz emd_16493.map.gz | 230.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16493-v30.xml emd-16493-v30.xml emd-16493.xml emd-16493.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16493.png emd_16493.png | 135.1 KB | ||
| Filedesc metadata |  emd-16493.cif.gz emd-16493.cif.gz | 7.1 KB | ||
| Others |  emd_16493_additional_1.map.gz emd_16493_additional_1.map.gz emd_16493_additional_2.map.gz emd_16493_additional_2.map.gz emd_16493_half_map_1.map.gz emd_16493_half_map_1.map.gz emd_16493_half_map_2.map.gz emd_16493_half_map_2.map.gz | 229.7 MB 229.7 MB 226.7 MB 226.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16493 http://ftp.pdbj.org/pub/emdb/structures/EMD-16493 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16493 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16493 | HTTPS FTP |
-Validation report
| Summary document |  emd_16493_validation.pdf.gz emd_16493_validation.pdf.gz | 1022.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16493_full_validation.pdf.gz emd_16493_full_validation.pdf.gz | 1022.3 KB | Display | |
| Data in XML |  emd_16493_validation.xml.gz emd_16493_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_16493_validation.cif.gz emd_16493_validation.cif.gz | 18.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16493 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16493 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16493 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16493 | HTTPS FTP |
-Related structure data
| Related structure data |  8c8tMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16493.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16493.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall cryo-EM map of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
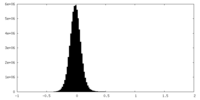
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local refined cryo-EM map of first bNAbs EPTC112...
| File | emd_16493_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refined cryo-EM map of first bNAbs EPTC112 bound to BG505 SOSIP.664 HIV-1 Env trimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
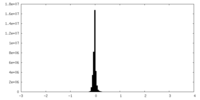
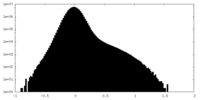
| Density Histograms |
-Additional map: Local refined cryo-EM map of second bNAbs EPTC112...
| File | emd_16493_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refined cryo-EM map of second bNAbs EPTC112 bound to BG505 SOSIP.664 HIV-1 Env trimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
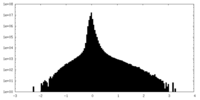
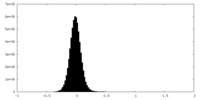
| Density Histograms |
-Half map: Half cryo-EM map A of BG505 SOSIP.664 HIV-1...
| File | emd_16493_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half cryo-EM map A of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | ||||||||||||
| Projections & Slices |
| ||||||||||||
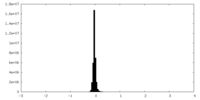
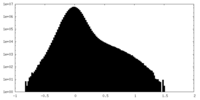
| Density Histograms |
-Half map: Half cryo-EM map B of BG505 SOSIP.664 HIV-1...
| File | emd_16493_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half cryo-EM map B of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | ||||||||||||
| Projections & Slices |
| ||||||||||||
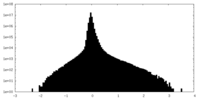
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of Bg505 Sosip.664 HIV-1 Env with bNAbs EPTC112 a...
| Entire | Name: Ternary complex of Bg505 Sosip.664 HIV-1 Env with bNAbs EPTC112 and 3BNC117 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Bg505 Sosip.664 HIV-1 Env with bNAbs EPTC112 a...
| Supramolecule | Name: Ternary complex of Bg505 Sosip.664 HIV-1 Env with bNAbs EPTC112 and 3BNC117 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Details: Gp120 Bg505 Sosip.664 T332n / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 52.5515 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LWVTVYYGVP VWKDAETTLF CASDAKAYET EKHNVWATHA CVPTDPNPQE IHLENVTEEF NMWKNNMVEQ MHTDIISLWD QSLKPCVKL TPLCVTLQCT NVTNNITDDM RGELKNCSFN MTTELRDKKQ KVYSLFYRLD VVQINENQGN RSNNSNKEYR L INCNTSAI ...String: LWVTVYYGVP VWKDAETTLF CASDAKAYET EKHNVWATHA CVPTDPNPQE IHLENVTEEF NMWKNNMVEQ MHTDIISLWD QSLKPCVKL TPLCVTLQCT NVTNNITDDM RGELKNCSFN MTTELRDKKQ KVYSLFYRLD VVQINENQGN RSNNSNKEYR L INCNTSAI TQACPKVSFE PIPIHYCAPA GFAILKCKDK KFNGTGPCPS VSTVQCTHGI KPVVSTQLLL NGSLAEEEVM IR SENITNN AKNILVQFNT PVQINCTRPN NNTRKSIRIG PGQAFYATGD IIGDIRQAHC NVSKATWNET LGKVVKQLRK HFG NNTIIR FANSSGGDLE VTTHSFNCGG EFFYCNTSGL FNSTWISNTS VQGSNSTGSN DSITLPCRIK QIINMWQRIG QAMY APPIQ GVIRCVSNIT GLILTRDGGS TNSTTETFRP GGGDMRDNWR SELYKYKVVK IEPLGVAPTR CKRR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Gp41 Bg505 T332n Sosip.664
| Macromolecule | Name: Gp41 Bg505 T332n Sosip.664 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 16.431627 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LGFLGAAGST MGAASMTLTV QARNLLSGIV QQQSNLLRAP EAQQHLLKLT VWGIKQLQAR VLAVERYLRD QQLLGIWGCS GKLICCTNV PWNSSWSNRN LSEIWDNMTW LQWDKEISNY TQIIYGLLEE SQNQQEKNEQ DLLALD |
-Macromolecule #3: IgG EPTC112 Fab fragment heavy chain
| Macromolecule | Name: IgG EPTC112 Fab fragment heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.320389 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLLESGPG LVRPSETLTL TCSVFNSRVS GYYYSWIRQP PGRGLEWIAS THFSLRPSRN PSLLSRVTTS IDTERYQVFL NMRSVTAAD TAVYFCARGD ASGWRADYFP HWGQGTLVVV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: QVQLLESGPG LVRPSETLTL TCSVFNSRVS GYYYSWIRQP PGRGLEWIAS THFSLRPSRN PSLLSRVTTS IDTERYQVFL NMRSVTAAD TAVYFCARGD ASGWRADYFP HWGQGTLVVV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKSCDKTH HHHHH |
-Macromolecule #4: IgG 3BNC117 Fab heavy chain
| Macromolecule | Name: IgG 3BNC117 Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.640484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLLQSGAA VTKPGASVRV SCEASGYNIR DYFIHWWRQA PGQGLQWVGW INPKTGQPNN PRQFQGRVSL TRHASWDFDT FSFYMDLKA LRSDDTAVYF CARQRSDYWD FDVWGSGTQV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QVQLLQSGAA VTKPGASVRV SCEASGYNIR DYFIHWWRQA PGQGLQWVGW INPKTGQPNN PRQFQGRVSL TRHASWDFDT FSFYMDLKA LRSDDTAVYF CARQRSDYWD FDVWGSGTQV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSC |
-Macromolecule #5: IgG 3BNC117 Fab Light Chain
| Macromolecule | Name: IgG 3BNC117 Fab Light Chain / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.022658 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDTVT ITCQANGYLN WYQQRRGKAP KLLIYDGSKL ERGVPSRFSG RRWGQEYNLT INNLQPEDIA TYFCQVYEF VVPGTRLDLK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG NSQESVTEQD S KDSTYSLS ...String: DIQMTQSPSS LSASVGDTVT ITCQANGYLN WYQQRRGKAP KLLIYDGSKL ERGVPSRFSG RRWGQEYNLT INNLQPEDIA TYFCQVYEF VVPGTRLDLK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG NSQESVTEQD S KDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #6: IgG EPTC112 Fab Light Chain
| Macromolecule | Name: IgG EPTC112 Fab Light Chain / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.680932 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSA SGSPGQSVTI SCTGTSSDIG ASDYVSWYQQ YPGEAPKVII YDVTKRPSGV PDRFSGSKSG TTASLTVSGL QAEDEADYY CSSDAGRHTL LFGGGTKVTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: QSVLTQPPSA SGSPGQSVTI SCTGTSSDIG ASDYVSWYQQ YPGEAPKVII YDVTKRPSGV PDRFSGSKSG TTASLTVSGL QAEDEADYY CSSDAGRHTL LFGGGTKVTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Macromolecule #10: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 10 / Number of copies: 24 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: PBS |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 251591 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)