+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Drosophila melanogaster Rab7 GEF complex Mon1-Ccz1-Bulli | |||||||||
 Map data Map data | density modified composite map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Guanain Nucleotide Exchange Factor / membrane traffic / lysosome / ENDOCYTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationguanyl nucleotide exchange factor inhibitor activity / Mon1-Ccz1 complex / RAB GEFs exchange GTP for GDP on RABs / guanyl-nucleotide exchange factor adaptor activity / protein targeting to vacuole / endosome to lysosome transport via multivesicular body sorting pathway / neuromuscular junction development / synaptic cleft / vesicle-mediated transport / guanyl-nucleotide exchange factor activity ...guanyl nucleotide exchange factor inhibitor activity / Mon1-Ccz1 complex / RAB GEFs exchange GTP for GDP on RABs / guanyl-nucleotide exchange factor adaptor activity / protein targeting to vacuole / endosome to lysosome transport via multivesicular body sorting pathway / neuromuscular junction development / synaptic cleft / vesicle-mediated transport / guanyl-nucleotide exchange factor activity / autophagy / late endosome membrane / regulation of autophagy / intracellular membrane-bounded organelle / lipid binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
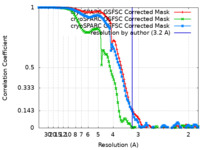
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Schaefer J / Herrmann E / Kuemmel D / Moeller A | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structure of the metazoan Rab7 GEF complex Mon1-Ccz1-Bulli. Authors: Eric Herrmann / Jan-Hannes Schäfer / Stephan Wilmes / Christian Ungermann / Arne Moeller / Daniel Kümmel /  Abstract: The endosomal system of eukaryotic cells represents a central sorting and recycling compartment linked to metabolic signaling and the regulation of cell growth. Tightly controlled activation of Rab ...The endosomal system of eukaryotic cells represents a central sorting and recycling compartment linked to metabolic signaling and the regulation of cell growth. Tightly controlled activation of Rab GTPases is required to establish the different domains of endosomes and lysosomes. In metazoans, Rab7 controls endosomal maturation, autophagy, and lysosomal function. It is activated by the guanine nucleotide exchange factor (GEF) complex Mon1-Ccz1-Bulli (MCBulli) of the tri-longin domain (TLD) family. While the Mon1 and Ccz1 subunits have been shown to constitute the active site of the complex, the role of Bulli remains elusive. We here present the cryo-electron microscopy (cryo-EM) structure of MCBulli at 3.2 Å resolution. Bulli associates as a leg-like extension at the periphery of the Mon1 and Ccz1 heterodimers, consistent with earlier reports that Bulli does not impact the activity of the complex or the interactions with recruiter and substrate GTPases. While MCBulli shows structural homology to the related ciliogenesis and planar cell polarity effector (Fuzzy-Inturned-Wdpcp) complex, the interaction of the TLD core subunits Mon1-Ccz1 and Fuzzy-Inturned with Bulli and Wdpcp, respectively, is remarkably different. The variations in the overall architecture suggest divergent functions of the Bulli and Wdpcp subunits. Based on our structural analysis, Bulli likely serves as a recruitment platform for additional regulators of endolysosomal trafficking to sites of Rab7 activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16457.map.gz emd_16457.map.gz | 106.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16457-v30.xml emd-16457-v30.xml emd-16457.xml emd-16457.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16457_fsc.xml emd_16457_fsc.xml emd_16457_fsc_2.xml emd_16457_fsc_2.xml emd_16457_fsc_3.xml emd_16457_fsc_3.xml | 10.6 KB 10.6 KB 10.6 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_16457.png emd_16457.png | 92.2 KB | ||
| Filedesc metadata |  emd-16457.cif.gz emd-16457.cif.gz | 7.1 KB | ||
| Others |  emd_16457_additional_1.map.gz emd_16457_additional_1.map.gz emd_16457_additional_2.map.gz emd_16457_additional_2.map.gz emd_16457_additional_3.map.gz emd_16457_additional_3.map.gz emd_16457_additional_4.map.gz emd_16457_additional_4.map.gz | 114.7 MB 118 MB 117.8 MB 118 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16457 http://ftp.pdbj.org/pub/emdb/structures/EMD-16457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16457 | HTTPS FTP |
-Related structure data
| Related structure data |  8c7gMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16457.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16457.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified composite map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.924 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: composite map of L1 and L2
| File | emd_16457_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map of L1 and L2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
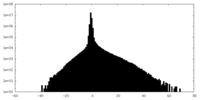
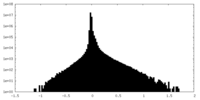
| Density Histograms |
-Additional map: consensus map
| File | emd_16457_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | consensus map | ||||||||||||
| Projections & Slices |
| ||||||||||||
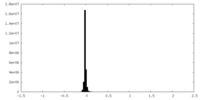
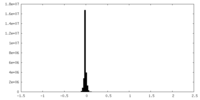
| Density Histograms |
-Additional map: local mal MCB-bulli (L2)
| File | emd_16457_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local mal MCB-bulli (L2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
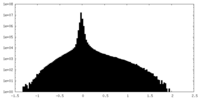
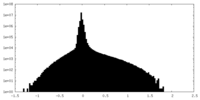
| Density Histograms |
-Additional map: local map MCB-core (L1)
| File | emd_16457_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local map MCB-core (L1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
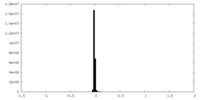
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric metazoan guanine-nucleotide-exchange factor Mon1-Ccz1-Bulli
| Entire | Name: Trimeric metazoan guanine-nucleotide-exchange factor Mon1-Ccz1-Bulli |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric metazoan guanine-nucleotide-exchange factor Mon1-Ccz1-Bulli
| Supramolecule | Name: Trimeric metazoan guanine-nucleotide-exchange factor Mon1-Ccz1-Bulli type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Mic1 domain-containing protein
| Macromolecule | Name: Mic1 domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.058391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDNSNGIHYI ELTPNPIRFD AVSQLTNVFF DDSNKQIFAV RSGGATGVVV KGPGSPDDVV ISFCMSDRGG AIRSIKFSPD NQILAVQRK ENSVEFICFQ GDQPLLQDII THQVKTLIHG FVWVHNREVA LISNTGVEVY TVVPEKRQVR SVKSLSIGIK W FAWCCDAN ...String: MDNSNGIHYI ELTPNPIRFD AVSQLTNVFF DDSNKQIFAV RSGGATGVVV KGPGSPDDVV ISFCMSDRGG AIRSIKFSPD NQILAVQRK ENSVEFICFQ GDQPLLQDII THQVKTLIHG FVWVHNREVA LISNTGVEVY TVVPEKRQVR SVKSLSIGIK W FAWCCDAN VALLCTSEGN SLIPVLVKQK VITKLPKVDL GNPSRDVQES KVTLGQVYGV LAVLILQSNS TTGLMEVEVH LL NGPGLAP RKCHVLRLSL LGRFAINTVD NLIVVHHQAS GTSLLFDISL PGEVINEITY HTPITPGRSI KPFGLKLPSL SPD GQILQC ELYSTHWVLF QPNIVIDAKL GCMWFLNLCI EPLCQLISDR IRLTEFLLQR SNGKQMLLKV IGQLVDDQYK GTLL PVLET IFSRINKIYA SWVQLELQNQ TAQPSNVKTT TLKQSTPPIV LIEQLDMVQI FQRIARRPYT ESILMLYLQS LNKFN IAAQ EELSKMIISE LISNRSFDTL RRLVSYSMLL ESKSVACFLL SHSNVDTAIS QVAIDMLGRI EAHEIIIEVM LGQGKV IDA LRLAKNSMGL EKVPARKFLE AAHKTKDDLI FHSVYRFFQM RNLKLYETLS FPKAEQCTEF IQHYNNTFPA DNPTRQP VS UniProtKB: Regulator of MON1-CCZ1 complex |
-Macromolecule #2: Caffeine, calcium, zinc sensitivity 1
| Macromolecule | Name: Caffeine, calcium, zinc sensitivity 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 58.308566 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKLLQRVEI TLRSFYIFNS TFGQVEGEEH KKVLFYHPND IELNTKIKDV GLSEAIIRFT GTFTSEDDCQ ALHTQKTTQL FYQPEPGYW LVLVLNVPKE VRLKEGVEVA DYRGAEISDR IYRAILRQCY QMFRFQNGCF SSCGSEEPNP DKRRELLCQK L LQFYDQHL ...String: MAKLLQRVEI TLRSFYIFNS TFGQVEGEEH KKVLFYHPND IELNTKIKDV GLSEAIIRFT GTFTSEDDCQ ALHTQKTTQL FYQPEPGYW LVLVLNVPKE VRLKEGVEVA DYRGAEISDR IYRAILRQCY QMFRFQNGCF SSCGSEEPNP DKRRELLCQK L LQFYDQHL TNLRDPAQCD IIDMLHSIQY LPLDKTLFLR AQNFGTLCET FPDIKESIML YQEQVLCGGK LSPEDLHCVH SY VVQHVLK VEASSSTIAV SPSLKRSISE CQVGGFVRSR QKVAGDEHDA VNEEDHPMKV YVTLDKEAKP YYLLIYRALH ITL CLFLNA DQVAPKQDLY DDLHAYMAPQ LTSLARDISS ELTKEAVGAA GQDNSSGNSE TAPKYLFINE QSLQHHTNFQ RHLP QGLPR NVLSIIADLA NGSGKAEMES APAEEVQVKT TNDYWIVKRR CNYRQYYVIL CNSKATLLDV TQEARRIFEQ ELTDD VFFD KDYKDHDGDY KDHDIDYKDD DDK UniProtKB: Vacuolar fusion protein CCZ1 homolog |
-Macromolecule #3: Vacuolar fusion protein MON1 homolog
| Macromolecule | Name: Vacuolar fusion protein MON1 homolog / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.904152 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEVEQTSVRS DTNSTCEYLD AEGDPESPNL YQEADPDQEA EQQNHSIISE LRDGLGTMRD NSALSPEPGQ ENKGLAASVE SLALSTSTS AKTEDSIGGG LEEEYDYQHD SLWQGQKKHI FILSEAGKPI FSLHGNEDKL ATLFGVIQAL VSFVQMGQDA I TSIHAGGI ...String: MEVEQTSVRS DTNSTCEYLD AEGDPESPNL YQEADPDQEA EQQNHSIISE LRDGLGTMRD NSALSPEPGQ ENKGLAASVE SLALSTSTS AKTEDSIGGG LEEEYDYQHD SLWQGQKKHI FILSEAGKPI FSLHGNEDKL ATLFGVIQAL VSFVQMGQDA I TSIHAGGI KFAFMQRSSL ILVAASRSNM SVQQLQLQLG DVYNQILSIL TYSHMTKIFE RRKNFDLRRL LSGSERLFYN LL ANDSSSA KVSNNIFTFL TNSIRVFPLP TTIRSQITSA IQSNCSKIKN LVFAVLIANN KLIALVRMKK YSIHPADLRL IFN LVECSE SFKSSENWSP ICLPKFDMNG YLHAHVSYLA DDCQACLLLL SVDRDAFFTL AEAKAKITEK LRKSHCLEAI NEEL QQPFN AKLYQQVVGI PELRHFLYKP KSTAQLLCPM LRHPYKSLTE LERLEAIYCD LLHRIHNSSR PLKLIYEMKE REVVL AWAT GTYELYAIFE PVVDKATVIK YVDKLIKWIE KEYDVYFIRN HATF UniProtKB: Vacuolar fusion protein MON1 homolog |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.3 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 5931 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 129 |
|---|---|
| Output model |  PDB-8c7g: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)