登録情報 データベース : EMDB / ID : EMD-16452タイトル Human spliceosomal PM5 C* complex map of the human spliceosomal PM5 C* complex 複合体 : human spliceosomal C* complex assembled on PM5 pre-mRNARNA : x 5種タンパク質・ペプチド : x 48種リガンド : x 6種 / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
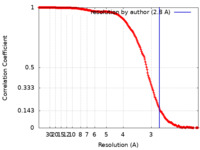
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 2.8 Å Dybkov O / Kastner B / Luehrmann R 資金援助 Organization Grant number 国 Max Planck Society
ジャーナル : Sci Adv / 年 : 2023タイトル : Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.著者: Olexandr Dybkov / Marco Preußner / Leyla El Ayoubi / Vivi-Yun Feng / Caroline Harnisch / Kilian Merz / Paula Leupold / Peter Yudichev / Dmitry E Agafonov / Cindy L Will / Cyrille Girard / ... 著者 : Olexandr Dybkov / Marco Preußner / Leyla El Ayoubi / Vivi-Yun Feng / Caroline Harnisch / Kilian Merz / Paula Leupold / Peter Yudichev / Dmitry E Agafonov / Cindy L Will / Cyrille Girard / Christian Dienemann / Henning Urlaub / Berthold Kastner / Florian Heyd / Reinhard Lührmann / 要旨 : Alternative precursor messenger RNA splicing is instrumental in expanding the proteome of higher eukaryotes, and changes in 3' splice site (3'ss) usage contribute to human disease. We demonstrate by ... Alternative precursor messenger RNA splicing is instrumental in expanding the proteome of higher eukaryotes, and changes in 3' splice site (3'ss) usage contribute to human disease. We demonstrate by small interfering RNA-mediated knockdowns, followed by RNA sequencing, that many proteins first recruited to human C* spliceosomes, which catalyze step 2 of splicing, regulate alternative splicing, including the selection of alternatively spliced NAGNAG 3'ss. Cryo-electron microscopy and protein cross-linking reveal the molecular architecture of these proteins in C* spliceosomes, providing mechanistic and structural insights into how they influence 3'ss usage. They further elucidate the path of the 3' region of the intron, allowing a structure-based model for how the C* spliceosome potentially scans for the proximal 3'ss. By combining biochemical and structural approaches with genome-wide functional analyses, our studies reveal widespread regulation of alternative 3'ss usage after step 1 of splicing and the likely mechanisms whereby C* proteins influence NAGNAG 3'ss choices. 履歴 登録 2023年1月12日 - ヘッダ(付随情報) 公開 2023年7月12日 - マップ公開 2023年7月12日 - 更新 2024年11月6日 - 現状 2024年11月6日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 ドイツ, 1件
ドイツ, 1件  引用
引用 ジャーナル: Sci Adv / 年: 2023
ジャーナル: Sci Adv / 年: 2023
 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_16452.map.gz
emd_16452.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-16452-v30.xml
emd-16452-v30.xml emd-16452.xml
emd-16452.xml EMDBヘッダ
EMDBヘッダ emd_16452_fsc.xml
emd_16452_fsc.xml FSCデータファイル
FSCデータファイル emd_16452.png
emd_16452.png emd-16452.cif.gz
emd-16452.cif.gz emd_16452_additional_1.map.gz
emd_16452_additional_1.map.gz emd_16452_half_map_1.map.gz
emd_16452_half_map_1.map.gz emd_16452_half_map_2.map.gz
emd_16452_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-16452
http://ftp.pdbj.org/pub/emdb/structures/EMD-16452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16452
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16452
 F&H 検索
F&H 検索 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_16452.map.gz / 形式: CCP4 / 大きさ: 744.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_16452.map.gz / 形式: CCP4 / 大きさ: 744.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析
 ムービー
ムービー コントローラー
コントローラー




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)