[English] 日本語
 Yorodumi
Yorodumi- EMDB-1641: Helical reconstruction of the bacterial L-type flagella filament -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1641 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of the bacterial L-type flagella filament | |||||||||
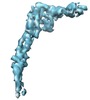
 Map data Map data | This is a cryo-EM map of L-type flagellin including data up to 4 Angstrom resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial flagellum | |||||||||
| Function / homology |  Function and homology information Function and homology informationTLR5 cascade / MyD88 cascade initiated on plasma membrane / NFkB and MAPK activation mediated by TRAF6 / The IPAF inflammasome / bacterial-type flagellum / receptor ligand activity / structural molecule activity / extracellular space Similarity search - Function | |||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Maki-Yonekura S / Yonekura K / Namba K | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2010 Journal: Nat Struct Mol Biol / Year: 2010Title: Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Authors: Saori Maki-Yonekura / Koji Yonekura / Keiichi Namba /  Abstract: The bacterial flagellar filament is a helical propeller rotated by the flagellar motor for bacterial locomotion. The filament is a supercoiled assembly of a single protein, flagellin, and is formed ...The bacterial flagellar filament is a helical propeller rotated by the flagellar motor for bacterial locomotion. The filament is a supercoiled assembly of a single protein, flagellin, and is formed by 11 protofilaments. For bacterial taxis, the reversal of motor rotation switches the supercoil between left- and right-handed, both of which arise from combinations of two distinct conformations and packing interactions of the L-type and R-type protofilaments. Here we report an atomic model of the L-type straight filament by electron cryomicroscopy and helical image analysis. Comparison with the R-type structure shows interesting features: an orientation change of the outer core domains (D1) against the inner core domains (D0) showing almost invariant orientation and packing, a conformational switching within domain D1, and the conformational flexibility of domains D0 and D1 with their spoke-like connection for tight molecular packing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1641.map.gz emd_1641.map.gz | 454.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1641-v30.xml emd-1641-v30.xml emd-1641.xml emd-1641.xml | 7.7 KB 7.7 KB | Display Display |  EMDB header EMDB header |
| Images |  em-1641.png em-1641.png | 170.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1641 http://ftp.pdbj.org/pub/emdb/structures/EMD-1641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1641 | HTTPS FTP |
-Related structure data
| Related structure data |  3a5xMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1641.map.gz / Format: CCP4 / Size: 32 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1641.map.gz / Format: CCP4 / Size: 32 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a cryo-EM map of L-type flagellin including data up to 4 Angstrom resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : S. enterica serovar Typhimurium L-type flagella filament
| Entire | Name: S. enterica serovar Typhimurium L-type flagella filament |
|---|---|
| Components |
|
-Supramolecule #1000: S. enterica serovar Typhimurium L-type flagella filament
| Supramolecule | Name: S. enterica serovar Typhimurium L-type flagella filament type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Macromolecule #1: Flagella filament
| Macromolecule | Name: Flagella filament / type: protein_or_peptide / ID: 1 / Name.synonym: Flagella filament / Oligomeric state: Helical assembly / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)Strain: SJW1660 synonym: Salmonella enterica subsp. enterica serovar Typhimurium Location in cell: Outside cell |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3000SFF |
|---|---|
| Temperature | Average: 4 K |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 5 µm |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Top entry / Specimen holder model: JEOL |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: OTHER / Software - Name: Handmade |
|---|---|
| CTF correction | Details: Each filament |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)