[English] 日本語
 Yorodumi
Yorodumi- EMDB-16280: Cryo-EM structure of rat SLC22A6 bound to alpha-ketoglutaric acid -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of rat SLC22A6 bound to alpha-ketoglutaric acid | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | membrane protein / transporter | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationOrganic anion transport by SLC22 transporters / renal tubular secretion / alpha-ketoglutarate transport / alpha-ketoglutarate transmembrane transporter activity / sodium-independent organic anion transport / : / metanephric proximal tubule development / prostaglandin transport / prostaglandin transmembrane transporter activity / solute:inorganic anion antiporter activity ...Organic anion transport by SLC22 transporters / renal tubular secretion / alpha-ketoglutarate transport / alpha-ketoglutarate transmembrane transporter activity / sodium-independent organic anion transport / : / metanephric proximal tubule development / prostaglandin transport / prostaglandin transmembrane transporter activity / solute:inorganic anion antiporter activity / organic anion transport / : / monoatomic anion transport / chloride ion binding / antiporter activity / xenobiotic transmembrane transporter activity / transmembrane transporter activity / basal plasma membrane / caveola / basolateral plasma membrane / protein-containing complex / identical protein binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
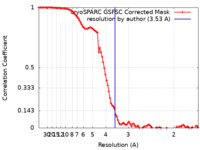
| Method | single particle reconstruction / cryo EM / Resolution: 3.53 Å | ||||||||||||
 Authors Authors | Parker JL / Kato T / Newstead S | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1. Authors: Joanne L Parker / Takafumi Kato / Gabriel Kuteyi / Oleg Sitsel / Simon Newstead /   Abstract: In mammals, the kidney plays an essential role in maintaining blood homeostasis through the selective uptake, retention or elimination of toxins, drugs and metabolites. Organic anion transporters ...In mammals, the kidney plays an essential role in maintaining blood homeostasis through the selective uptake, retention or elimination of toxins, drugs and metabolites. Organic anion transporters (OATs) are responsible for the recognition of metabolites and toxins in the nephron and their eventual urinary excretion. Inhibition of OATs is used therapeutically to improve drug efficacy and reduce nephrotoxicity. The founding member of the renal organic anion transporter family, OAT1 (also known as SLC22A6), uses the export of α-ketoglutarate (α-KG), a key intermediate in the Krebs cycle, to drive selective transport and is allosterically regulated by intracellular chloride. However, the mechanisms linking metabolite cycling, drug transport and intracellular chloride remain obscure. Here, we present cryogenic-electron microscopy structures of OAT1 bound to α-KG, the antiviral tenofovir and clinical inhibitor probenecid, used in the treatment of Gout. Complementary in vivo cellular assays explain the molecular basis for α-KG driven drug elimination and the allosteric regulation of organic anion transport in the kidney by chloride. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16280.map.gz emd_16280.map.gz | 85.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16280-v30.xml emd-16280-v30.xml emd-16280.xml emd-16280.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16280_fsc.xml emd_16280_fsc.xml | 9.6 KB | Display |  FSC data file FSC data file |
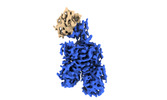
| Images |  emd_16280.png emd_16280.png | 40.8 KB | ||
| Masks |  emd_16280_msk_1.map emd_16280_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16280.cif.gz emd-16280.cif.gz | 6.2 KB | ||
| Others |  emd_16280_half_map_1.map.gz emd_16280_half_map_1.map.gz emd_16280_half_map_2.map.gz emd_16280_half_map_2.map.gz | 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16280 http://ftp.pdbj.org/pub/emdb/structures/EMD-16280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16280 | HTTPS FTP |
-Related structure data
| Related structure data |  8bw7MC  8bvrC  8bvsC  8bvtC  8omuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16280.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16280.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
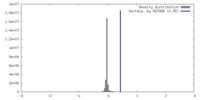
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16280_msk_1.map emd_16280_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
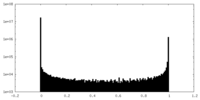
| Density Histograms |
-Half map: #2
| File | emd_16280_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
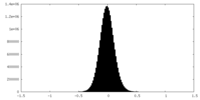
| Density Histograms |
-Half map: #1
| File | emd_16280_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
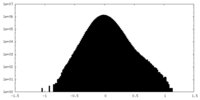
| Density Histograms |
- Sample components
Sample components
-Entire : OAT1
| Entire | Name: OAT1 |
|---|---|
| Components |
|
-Supramolecule #1: OAT1
| Supramolecule | Name: OAT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Solute carrier family 22 member 6
| Macromolecule | Name: Solute carrier family 22 member 6 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.553559 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAFNDLLKQV GGVGRFQLIQ VTMVVAPLLL MASHNTLQNF TAAIPPHHCR PPANANLSKD GGLEAWLPLD KQGQPESCLR FTSPQWGPP FYNGTEANGT RVTEPCIDGW VYDNSTFPST IVTEWNLVCS HRAFRQLAQS LYMVGVLLGA MVFGYLADRL G RRKVLILN ...String: MAFNDLLKQV GGVGRFQLIQ VTMVVAPLLL MASHNTLQNF TAAIPPHHCR PPANANLSKD GGLEAWLPLD KQGQPESCLR FTSPQWGPP FYNGTEANGT RVTEPCIDGW VYDNSTFPST IVTEWNLVCS HRAFRQLAQS LYMVGVLLGA MVFGYLADRL G RRKVLILN YLQTAVSGTC AAYAPNYTVY CVFRLLSGMS LASIAINCMT LNVEWMPIHT RAYVGTLIGY VYSLGQFLLA GI AYAVPHW RHLQLVVSVP FFIAFIYSWF FIESARWYSS SGRLDLTLRA LQRVARINGK QEEGAKLSIE VLRTSLQKEL TLS KGQASA MELLRCPTLR HLFLCLSMLW FATSFAYYGL VMDLQGFGVS MYLIQVIFGA VDLPAKFVCF LVINSMGRRP AQMA SLLLA GICILVNGII PKSHTIIRTS LAVLGKGCLA SSFNCIFLYT GELYPTVIRQ TGLGMGSTMA RVGSIVSPLV SMTAE FYPS MPLFIFGAVP VVASAVTALL PETLGQPLPD TVQDLKSRSR GKQNQQQQEQ QKQMMPLENL YFQ UniProtKB: Solute carrier family 22 member 6 |
-Macromolecule #2: Synthetic nanobody (Sybody)
| Macromolecule | Name: Synthetic nanobody (Sybody) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 16.079819 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGSSSQVQL VESGGGLVQA GGSLRLSCAA SGFPVKTEWM EWYRQAPGKE REWVAAIWSY GSGTRYADSV KGRFTISRDN AKNTVYLQM NSLKPEDTAV YYCLVRVGSW YHGQGTQVTV SAGRAGEQKL ISEEDLNSAV DHHHHHH |
-Macromolecule #3: 2-OXOGLUTARIC ACID
| Macromolecule | Name: 2-OXOGLUTARIC ACID / type: ligand / ID: 3 / Number of copies: 1 / Formula: AKG |
|---|---|
| Molecular weight | Theoretical: 146.098 Da |
| Chemical component information |  ChemComp-AKG: |
-Macromolecule #4: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8bw7: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)