+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | VaPomAB full length Saposin Nanodisc | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Flagellar sodium-driven stator unit / MOTOR PROTEIN | ||||||||||||
| Biological species |  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) | ||||||||||||
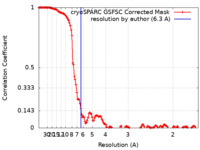
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | ||||||||||||
 Authors Authors | Hu H / Taylor NMI | ||||||||||||
| Funding support |  Denmark, 3 items Denmark, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Ion selectivity and rotor coupling of the Vibrio flagellar sodium-driven stator unit. Authors: Haidai Hu / Philipp F Popp / Mònica Santiveri / Aritz Roa-Eguiara / Yumeng Yan / Freddie J O Martin / Zheyi Liu / Navish Wadhwa / Yong Wang / Marc Erhardt / Nicholas M I Taylor /     Abstract: Bacteria swim using a flagellar motor that is powered by stator units. Vibrio spp. are highly motile bacteria responsible for various human diseases, the polar flagella of which are exclusively ...Bacteria swim using a flagellar motor that is powered by stator units. Vibrio spp. are highly motile bacteria responsible for various human diseases, the polar flagella of which are exclusively driven by sodium-dependent stator units (PomAB). However, how ion selectivity is attained, how ion transport triggers the directional rotation of the stator unit, and how the stator unit is incorporated into the flagellar rotor remained largely unclear. Here, we have determined by cryo-electron microscopy the structure of Vibrio PomAB. The electrostatic potential map uncovers sodium binding sites, which together with functional experiments and molecular dynamics simulations, reveal a mechanism for ion translocation and selectivity. Bulky hydrophobic residues from PomA prime PomA for clockwise rotation. We propose that a dynamic helical motif in PomA regulates the distance between PomA subunit cytoplasmic domains, stator unit activation, and torque transmission. Together, our study provides mechanistic insights for understanding ion selectivity and rotor incorporation of the stator unit of the bacterial flagellum. | ||||||||||||
| History |
|
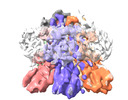
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16214.map.gz emd_16214.map.gz | 229.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16214-v30.xml emd-16214-v30.xml emd-16214.xml emd-16214.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16214_fsc.xml emd_16214_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_16214.png emd_16214.png | 112.8 KB | ||
| Filedesc metadata |  emd-16214.cif.gz emd-16214.cif.gz | 4.8 KB | ||
| Others |  emd_16214_half_map_1.map.gz emd_16214_half_map_1.map.gz emd_16214_half_map_2.map.gz emd_16214_half_map_2.map.gz | 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16214 http://ftp.pdbj.org/pub/emdb/structures/EMD-16214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16214 | HTTPS FTP |
-Validation report
| Summary document |  emd_16214_validation.pdf.gz emd_16214_validation.pdf.gz | 1000.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16214_full_validation.pdf.gz emd_16214_full_validation.pdf.gz | 1000.1 KB | Display | |
| Data in XML |  emd_16214_validation.xml.gz emd_16214_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_16214_validation.cif.gz emd_16214_validation.cif.gz | 27.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16214 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16214 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16214 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16214 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16214.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16214.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
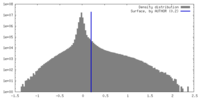
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16214_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
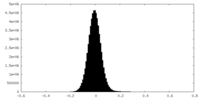
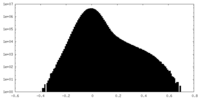
| Density Histograms |
-Half map: #2
| File | emd_16214_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
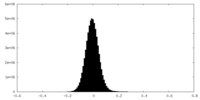
| Density Histograms |
- Sample components
Sample components
-Entire : flagellar sodium-driven stator unit
| Entire | Name: flagellar sodium-driven stator unit |
|---|---|
| Components |
|
-Supramolecule #1: flagellar sodium-driven stator unit
| Supramolecule | Name: flagellar sodium-driven stator unit / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
-Macromolecule #1: VaPomA
| Macromolecule | Name: VaPomA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Sequence | String: MDLATLLGLI GGFAFVIMAM VLGGSIGMFV DVTSILIVVG GSIFVVLMKF TMGQFFGATK IAGKAFMFKA DEPEDLIAKI VEMADAARKG GFLALEEMEI NNTFMQKGID LLVDGHDADV VRAALKKDIA LTDERHTQGT GVFRAFGDVA PAMGMIGTLV GLVAMLSNMD ...String: MDLATLLGLI GGFAFVIMAM VLGGSIGMFV DVTSILIVVG GSIFVVLMKF TMGQFFGATK IAGKAFMFKA DEPEDLIAKI VEMADAARKG GFLALEEMEI NNTFMQKGID LLVDGHDADV VRAALKKDIA LTDERHTQGT GVFRAFGDVA PAMGMIGTLV GLVAMLSNMD DPKAIGPAMA VALLTTLYGA ILSNMVFFPI ADKLSLRRDQ ETLNRRLIMD GVLAIQDGQN PRVIDSYLKN YLNEGKRALE IDE |
-Macromolecule #2: VaPoMB
| Macromolecule | Name: VaPoMB / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Sequence | String: MDDEDNKCDC PPPGLPLWMG TFADLMSLLM CFFVLLLSFS EMDVLKFKQI AGSMKFAFGV QNQLEVKDIP KGTSIIAQEF RPGRPEPTPI DVIMQENLYF QSQTMDITQQ TLEFHEGESE RAGGTKRDEG KLTGGQSPET STQNNESAEA DMQQQQSKEM SQEMETLMES ...String: MDDEDNKCDC PPPGLPLWMG TFADLMSLLM CFFVLLLSFS EMDVLKFKQI AGSMKFAFGV QNQLEVKDIP KGTSIIAQEF RPGRPEPTPI DVIMQENLYF QSQTMDITQQ TLEFHEGESE RAGGTKRDEG KLTGGQSPET STQNNESAEA DMQQQQSKEM SQEMETLMES IKKALEREIE QGAIEVENLG QQIVIRMREK GAFPEGSAFL QPKFRPLVRQ IAELVKDVPG IVRVSGHTDN RPLDSELYRS NWDLSSQRAV SVAQEMEKVR GFSHQRLRVR GMADTEPLLP NDSDENRALN RRVEISIMQG EPLYSEEVPV IQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 16 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 300mM NaCl, 20mM HEPES 7.5, 0.002%LMNG |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)