[English] 日本語
 Yorodumi
Yorodumi- EMDB-16169: Alpha7-nAChR extracellular ligand-binding domain (alpha7-ECD)in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alpha7-nAChR extracellular ligand-binding domain (alpha7-ECD)in complex with alpha-bungarotoxin. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Naja kaouthia (monocled cobra) Naja kaouthia (monocled cobra) | |||||||||
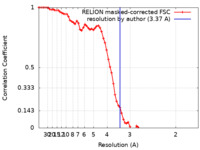
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Chesnokov YM / Kamyshinsky RA | |||||||||
| Funding support |  Russian Federation, 1 items Russian Federation, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: Membrane-mediated interaction of non-conventional snake three-finger toxins with nicotinic acetylcholine receptors. Authors: Zakhar O Shenkarev / Yuri M Chesnokov / Maxim M Zaigraev / Anton O Chugunov / Dmitrii S Kulbatskii / Milita V Kocharovskaya / Alexander S Paramonov / Maxim L Bychkov / Mikhail A Shulepko / ...Authors: Zakhar O Shenkarev / Yuri M Chesnokov / Maxim M Zaigraev / Anton O Chugunov / Dmitrii S Kulbatskii / Milita V Kocharovskaya / Alexander S Paramonov / Maxim L Bychkov / Mikhail A Shulepko / Dmitry E Nolde / Roman A Kamyshinsky / Evgeniy O Yablokov / Alexey S Ivanov / Mikhail P Kirpichnikov / Ekaterina N Lyukmanova /  Abstract: Nicotinic acetylcholine receptor of α7 type (α7-nAChR) presented in the nervous and immune systems and epithelium is a promising therapeutic target for cognitive disfunctions and cancer treatment. ...Nicotinic acetylcholine receptor of α7 type (α7-nAChR) presented in the nervous and immune systems and epithelium is a promising therapeutic target for cognitive disfunctions and cancer treatment. Weak toxin from Naja kaouthia venom (WTX) is a non-conventional three-finger neurotoxin, targeting α7-nAChR with weak affinity. There are no data on interaction mode of non-conventional neurotoxins with nAChRs. Using α-bungarotoxin (classical three-finger neurotoxin with high affinity to α7-nAChR), we showed applicability of cryo-EM to study complexes of α7-nAChR extracellular ligand-binding domain (α7-ECD) with toxins. Using cryo-EM structure of the α7-ECD/WTX complex, together with NMR data on membrane active site in the WTX molecule and mutagenesis data, we reconstruct the structure of α7-nAChR/WTX complex in the membrane environment. WTX interacts at the entrance to the orthosteric site located at the receptor intersubunit interface and simultaneously forms the contacts with the membrane surface. WTX interaction mode with α7-nAChR significantly differs from α-bungarotoxin's one, which does not contact the membrane. Our study reveals the important role of the membrane for interaction of non-conventional neurotoxins with the nicotinic receptors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16169.map.gz emd_16169.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16169-v30.xml emd-16169-v30.xml emd-16169.xml emd-16169.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16169_fsc.xml emd_16169_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16169.png emd_16169.png | 164 KB | ||
| Masks |  emd_16169_msk_1.map emd_16169_msk_1.map | 27 MB |  Mask map Mask map | |
| Others |  emd_16169_half_map_1.map.gz emd_16169_half_map_1.map.gz emd_16169_half_map_2.map.gz emd_16169_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16169 http://ftp.pdbj.org/pub/emdb/structures/EMD-16169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16169 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16169.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16169.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
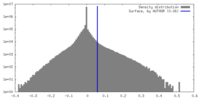
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16169_msk_1.map emd_16169_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16169_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16169_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Alpha7-nAChR (Nicotinic acetylcholine receptor of alpha7 type) ex...
| Entire | Name: Alpha7-nAChR (Nicotinic acetylcholine receptor of alpha7 type) extracellular ligand-binding domain |
|---|---|
| Components |
|
-Supramolecule #1: Alpha7-nAChR (Nicotinic acetylcholine receptor of alpha7 type) ex...
| Supramolecule | Name: Alpha7-nAChR (Nicotinic acetylcholine receptor of alpha7 type) extracellular ligand-binding domain type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10 KDa |
-Supramolecule #2: alpha-bungarotoxin
| Supramolecule | Name: alpha-bungarotoxin / type: complex / ID: 2 / Chimera: Yes / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Naja kaouthia (monocled cobra) Naja kaouthia (monocled cobra) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 5 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026000000000000002 kPa / Details: 15 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Cs corrector |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2245 / Average exposure time: 2.0 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.6 µm / Calibrated magnification: 75000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | PDB 4HQP was used to rigid body fit |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)