[English] 日本語
 Yorodumi
Yorodumi- EMDB-16134: Escherichia coli anaerobic fatty acid beta oxidation trifunctiona... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) octameric complex | ||||||||||||
 Map data Map data | AnEcTFE octamer | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | complex / heterooctamer / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationfatty acid beta-oxidation, unsaturated, even number / fatty acid beta-oxidation multienzyme complex / 3-hydroxybutyryl-CoA epimerase / 3-hydroxybutyryl-CoA epimerase activity / acetyl-CoA C-acyltransferase / long-chain (3S)-3-hydroxyacyl-CoA dehydrogenase (NAD+) activity / acetyl-CoA C-acyltransferase activity / 3-hydroxyacyl-CoA dehydrogenase / enoyl-CoA hydratase / (3S)-3-hydroxyacyl-CoA dehydrogenase (NAD+) activity ...fatty acid beta-oxidation, unsaturated, even number / fatty acid beta-oxidation multienzyme complex / 3-hydroxybutyryl-CoA epimerase / 3-hydroxybutyryl-CoA epimerase activity / acetyl-CoA C-acyltransferase / long-chain (3S)-3-hydroxyacyl-CoA dehydrogenase (NAD+) activity / acetyl-CoA C-acyltransferase activity / 3-hydroxyacyl-CoA dehydrogenase / enoyl-CoA hydratase / (3S)-3-hydroxyacyl-CoA dehydrogenase (NAD+) activity / acetyl-CoA C-acetyltransferase activity / enoyl-CoA hydratase activity / fatty acid beta-oxidation / NAD+ binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.3 Å | ||||||||||||
 Authors Authors | Sah-Teli SK / Pinkas M / Novacek J / Venkatesan R | ||||||||||||
| Funding support |  Czech Republic, Czech Republic,  Finland, European Union, 3 items Finland, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial β-oxidation trifunctional enzymes. Authors: Shiv K Sah-Teli / Matyas Pinkas / Mikko J Hynönen / Sarah J Butcher / Rik K Wierenga / Jiri Novacek / Rajaram Venkatesan /   Abstract: Facultative anaerobic bacteria such as Escherichia coli have two αβ heterotetrameric trifunctional enzymes (TFE), catalyzing the last three steps of the β-oxidation cycle: soluble aerobic TFE ...Facultative anaerobic bacteria such as Escherichia coli have two αβ heterotetrameric trifunctional enzymes (TFE), catalyzing the last three steps of the β-oxidation cycle: soluble aerobic TFE (EcTFE) and membrane-associated anaerobic TFE (anEcTFE), closely related to the human mitochondrial TFE (HsTFE). The cryo-EM structure of anEcTFE and crystal structures of anEcTFE-α show that the overall assembly of anEcTFE and HsTFE is similar. However, their membrane-binding properties differ considerably. The shorter A5-H7 and H8 regions of anEcTFE-α result in weaker α-β as well as α-membrane interactions, respectively. The protruding H-H region of anEcTFE-β is therefore more critical for membrane-association. Mutational studies also show that this region is important for the stability of the anEcTFE-β dimer and anEcTFE heterotetramer. The fatty acyl tail binding tunnel of the anEcTFE-α hydratase domain, as in HsTFE-α, is wider than in EcTFE-α, accommodating longer fatty acyl tails, in good agreement with their respective substrate specificities. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16134.map.gz emd_16134.map.gz | 14.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16134-v30.xml emd-16134-v30.xml emd-16134.xml emd-16134.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16134_fsc.xml emd_16134_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16134.png emd_16134.png | 76.3 KB | ||
| Filedesc metadata |  emd-16134.cif.gz emd-16134.cif.gz | 6.3 KB | ||
| Others |  emd_16134_half_map_1.map.gz emd_16134_half_map_1.map.gz emd_16134_half_map_2.map.gz emd_16134_half_map_2.map.gz | 164.9 MB 164.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16134 http://ftp.pdbj.org/pub/emdb/structures/EMD-16134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16134 | HTTPS FTP |
-Validation report
| Summary document |  emd_16134_validation.pdf.gz emd_16134_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16134_full_validation.pdf.gz emd_16134_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_16134_validation.xml.gz emd_16134_validation.xml.gz | 20.3 KB | Display | |
| Data in CIF |  emd_16134_validation.cif.gz emd_16134_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16134 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16134 | HTTPS FTP |
-Related structure data
| Related structure data |  8bnrMC  8bnuC  8brjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16134.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16134.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AnEcTFE octamer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
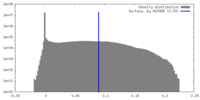
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: AnEcTFE octamer half2 map
| File | emd_16134_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AnEcTFE octamer half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
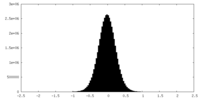
| Density Histograms |
-Half map: AnEcTFE octamer half1 map
| File | emd_16134_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AnEcTFE octamer half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
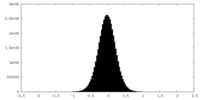
| Density Histograms |
- Sample components
Sample components
-Entire : anEcTFE octamer
| Entire | Name: anEcTFE octamer |
|---|---|
| Components |
|
-Supramolecule #1: anEcTFE octamer
| Supramolecule | Name: anEcTFE octamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: heterooctamer complex |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 3-ketoacyl-CoA thiolase FadI
| Macromolecule | Name: 3-ketoacyl-CoA thiolase FadI / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: acetyl-CoA C-acyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.20207 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQDPMGQVLP LVTRQGDRIA IVSGLRTPFA RQATAFHGIP AVDLGKMVVG ELLARSEIPA EVIEQLVFGQ VVQMPEAPN IAREIVLGTG MNVHTDAYSV SRACATSFQA VANVAESLMA GTIRAGIAGG ADSSSVLPIG VSKKLARVLV D VNKARTMS ...String: MGSSHHHHHH SQDPMGQVLP LVTRQGDRIA IVSGLRTPFA RQATAFHGIP AVDLGKMVVG ELLARSEIPA EVIEQLVFGQ VVQMPEAPN IAREIVLGTG MNVHTDAYSV SRACATSFQA VANVAESLMA GTIRAGIAGG ADSSSVLPIG VSKKLARVLV D VNKARTMS QRLKLFSRLR LRDLMPVPPA VAEYSTGLRM GDTAEQMAKT YGITREQQDA LAHRSHQRAA QAWSDGKLKE EV MTAFIPP YKQPLVEDNN IRGNSSLADY AKLRPAFDRK HGTVTAANST PLTDGAAAVI LMTESRAKEL GLVPLGYLRS YAF TAIDVW QDMLLGPAWS TPLALERAGL TMSDLTLIDM HEAFAAQTLA NIQLLGSERF AREALGRAHA TGEVDDSKFN VLGG SIAYG HPFAATGARM ITQTLHELRR RGGGFGLVTA CAAGGLGAAM VLEAE UniProtKB: 3-ketoacyl-CoA thiolase FadI |
-Macromolecule #2: Fatty acid oxidation complex subunit alpha
| Macromolecule | Name: Fatty acid oxidation complex subunit alpha / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: enoyl-CoA hydratase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 77.169125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEMTSAFTLN VRLDNIAVIT IDVPGEKMNT LKAEFASQVR AIIKQLRENK ELRGVVFVSA KPDNFIAGAD INMIGNCKTA QEAEALARQ GQQLMAEIHA LPIQVIAAIH GACLGGGLEL ALACHGRVCT DDPKTVLGLP EVQLGLLPGS GGTQRLPRLI G VSTALEMI ...String: MEMTSAFTLN VRLDNIAVIT IDVPGEKMNT LKAEFASQVR AIIKQLRENK ELRGVVFVSA KPDNFIAGAD INMIGNCKTA QEAEALARQ GQQLMAEIHA LPIQVIAAIH GACLGGGLEL ALACHGRVCT DDPKTVLGLP EVQLGLLPGS GGTQRLPRLI G VSTALEMI LTGKQLRAKQ ALKLGLVDDV VPHSILLEAA VELAKKERPS SRPLPVRERI LAGPLGRALL FKMVGKKTEH KT QGNYPAT ERILEVVETG LAQGTSSGYD AEARAFGELA MTPQSQALRS IFFASTDVKK DPGSDAPPAP LNSVGILGGG LMG GGIAYV TACKAGIPVR IKDINPQGIN HALKYSWDQL EGKVRRRHLK ASERDKQLAL ISGTTDYRGF AHRDLIIEAV FENL ELKQQ MVAEVEQNCA AHTIFASNTS SLPIGDIAAH ATRPEQVIGL HFFSPVEKMP LVEIIPHAGT SAQTIATTVK LAKKQ GKTP IVVRDKAGFY VNRILAPYIN EAIRMLTQGE RVEHIDAALV KFGFPVGPIQ LLDEVGIDTG TKIIPVLEAA YGERFS APA NVVSSILNDD RKGRKNGRGF YLYGQKGRKS KKQVDPAIYP LIGTQGQGRI SAPQVAERCV MLMLNEAVRC VDEQVIR SV RDGDIGAVFG IGFPPFLGGP FRYIDSLGAG EVVAIMQRLA TQYGSRFTPC ERLVEMGARG ESFWKTTATD LQ UniProtKB: Fatty acid oxidation complex subunit alpha |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 50 mM Tris, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, 0.0033% amphipol A8-35, pH 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Target criteria: cross-correlation coefficient |
|---|---|
| Output model |  PDB-8bnr: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)