+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical shell of CCMV capsid protein on DNA origami 6HB-2k | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | origami / capsid / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Bromovirus coat protein / Bromovirus coat protein / viral capsid / structural molecule activity / Coat protein Function and homology information Function and homology information | |||||||||
| Biological species |  Cowpea chlorotic mottle virus Cowpea chlorotic mottle virus | |||||||||
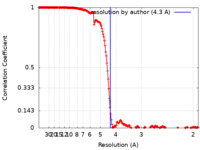
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Kumpula E-P / Seitz I / Kostiainen MA / Huiskonen JT | |||||||||
| Funding support | European Union,  Finland, 2 items Finland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Nanotechnol / Year: 2023 Journal: Nat Nanotechnol / Year: 2023Title: DNA-origami-directed virus capsid polymorphism. Authors: Iris Seitz / Sharon Saarinen / Esa-Pekka Kumpula / Donna McNeale / Eduardo Anaya-Plaza / Vili Lampinen / Vesa P Hytönen / Frank Sainsbury / Jeroen J L M Cornelissen / Veikko Linko / Juha T ...Authors: Iris Seitz / Sharon Saarinen / Esa-Pekka Kumpula / Donna McNeale / Eduardo Anaya-Plaza / Vili Lampinen / Vesa P Hytönen / Frank Sainsbury / Jeroen J L M Cornelissen / Veikko Linko / Juha T Huiskonen / Mauri A Kostiainen /     Abstract: Viral capsids can adopt various geometries, most iconically characterized by icosahedral or helical symmetries. Importantly, precise control over the size and shape of virus capsids would have ...Viral capsids can adopt various geometries, most iconically characterized by icosahedral or helical symmetries. Importantly, precise control over the size and shape of virus capsids would have advantages in the development of new vaccines and delivery systems. However, current tools to direct the assembly process in a programmable manner are exceedingly elusive. Here we introduce a modular approach by demonstrating DNA-origami-directed polymorphism of single-protein subunit capsids. We achieve control over the capsid shape, size and topology by employing user-defined DNA origami nanostructures as binding and assembly platforms, which are efficiently encapsulated within the capsid. Furthermore, the obtained viral capsid coatings can shield the encapsulated DNA origami from degradation. Our approach is, moreover, not limited to a single type of capsomers and can also be applied to RNA-DNA origami structures to pave way for next-generation cargo protection and targeting strategies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16076.map.gz emd_16076.map.gz | 375.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16076-v30.xml emd-16076-v30.xml emd-16076.xml emd-16076.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16076_fsc.xml emd_16076_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16076.png emd_16076.png | 165.8 KB | ||
| Masks |  emd_16076_msk_1.map emd_16076_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16076.cif.gz emd-16076.cif.gz | 5 KB | ||
| Others |  emd_16076_half_map_1.map.gz emd_16076_half_map_1.map.gz emd_16076_half_map_2.map.gz emd_16076_half_map_2.map.gz | 475.6 MB 475.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16076 http://ftp.pdbj.org/pub/emdb/structures/EMD-16076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16076 | HTTPS FTP |
-Validation report
| Summary document |  emd_16076_validation.pdf.gz emd_16076_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16076_full_validation.pdf.gz emd_16076_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_16076_validation.xml.gz emd_16076_validation.xml.gz | 26.4 KB | Display | |
| Data in CIF |  emd_16076_validation.cif.gz emd_16076_validation.cif.gz | 34.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16076 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16076 | HTTPS FTP |
-Related structure data
| Related structure data |  8bi4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16076.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16076.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16076_msk_1.map emd_16076_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16076_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16076_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helical shell of CCMV capsid protein on DNA origami 6HB-2k
| Entire | Name: Helical shell of CCMV capsid protein on DNA origami 6HB-2k |
|---|---|
| Components |
|
-Supramolecule #1: Helical shell of CCMV capsid protein on DNA origami 6HB-2k
| Supramolecule | Name: Helical shell of CCMV capsid protein on DNA origami 6HB-2k type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Cowpea chlorotic mottle virus Cowpea chlorotic mottle virus |
-Macromolecule #1: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cowpea chlorotic mottle virus Cowpea chlorotic mottle virus |
| Molecular weight | Theoretical: 20.336252 KDa |
| Sequence | String: MSTVGTGKLT RAQRRAAARK NKRNTRVVQP VIVEPIASGQ GKAIKAWTGY SVSKWTASCA AAEAKVTSAI TISLPNELSS ERNKQLKVG RVLLWLGLLP SVSGTVKSCV TETQTTAAAS FQVALAVADN SKDVVAAMYP EAFKGITLEQ LAADLTIYLY S SAALTEGD VIVHLEVEHV RPTFDDSFTP VY UniProtKB: Coat protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial model pdb:1cwp was first manually adjusted to density in ISOLDE and then refined in PHENIX |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8bi4: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)