+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | ISDra2 TnpB in complex with reRNA | |||||||||||||||
 Map data Map data | Sharpened map used for model building. Output of phenix.auto_refine v1.20.1-4487 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Transposon / TnpB / reRNA / RNA BINDING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtransposition / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / endonuclease activity / DNA recombination / DNA binding / RNA binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Deinococcus radiodurans (radioresistant) / Deinococcus radiodurans (radioresistant) /  Deinococcus radiodurans R1 (radioresistant) Deinococcus radiodurans R1 (radioresistant) | |||||||||||||||
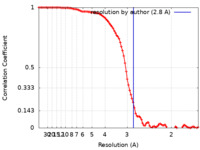
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||
 Authors Authors | Sasnauskas G / Tamulaitiene G / Carabias A / Siksnys V / Montoya G / Druteika G / Silanskas A / Venclovas C / Karvelis T / Kazlauskas D | |||||||||||||||
| Funding support |  Denmark, 4 items Denmark, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: TnpB structure reveals minimal functional core of Cas12 nuclease family. Authors: Giedrius Sasnauskas / Giedre Tamulaitiene / Gytis Druteika / Arturo Carabias / Arunas Silanskas / Darius Kazlauskas / Česlovas Venclovas / Guillermo Montoya / Tautvydas Karvelis / Virginijus Siksnys /  Abstract: The widespread TnpB proteins of IS200/IS605 transposon family have recently emerged as the smallest RNA-guided nucleases capable of targeted genome editing in eukaryotic cells. Bioinformatic analysis ...The widespread TnpB proteins of IS200/IS605 transposon family have recently emerged as the smallest RNA-guided nucleases capable of targeted genome editing in eukaryotic cells. Bioinformatic analysis identified TnpB proteins as the likely predecessors of Cas12 nucleases, which along with Cas9 are widely used for targeted genome manipulation. Whereas Cas12 family nucleases are well characterized both biochemically and structurally, the molecular mechanism of TnpB remains unknown. Here we present the cryogenic-electron microscopy structures of the Deinococcus radiodurans TnpB-reRNA (right-end transposon element-derived RNA) complex in DNA-bound and -free forms. The structures reveal the basic architecture of TnpB nuclease and the molecular mechanism for DNA target recognition and cleavage that is supported by biochemical experiments. Collectively, these results demonstrate that TnpB represents the minimal structural and functional core of the Cas12 protein family and provide a framework for developing TnpB-based genome editing tools. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16016.map.gz emd_16016.map.gz | 67.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16016-v30.xml emd-16016-v30.xml emd-16016.xml emd-16016.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16016_fsc.xml emd_16016_fsc.xml | 12.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_16016.png emd_16016.png | 161.1 KB | ||
| Masks |  emd_16016_msk_1.map emd_16016_msk_1.map | 75.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16016.cif.gz emd-16016.cif.gz | 6.5 KB | ||
| Others |  emd_16016_additional_1.map.gz emd_16016_additional_1.map.gz emd_16016_half_map_1.map.gz emd_16016_half_map_1.map.gz emd_16016_half_map_2.map.gz emd_16016_half_map_2.map.gz | 37.4 MB 69.6 MB 69.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16016 http://ftp.pdbj.org/pub/emdb/structures/EMD-16016 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16016 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16016 | HTTPS FTP |
-Related structure data
| Related structure data |  8bf8MC  8ex9C  8exaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16016.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16016.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map used for model building. Output of phenix.auto_refine v1.20.1-4487 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16016_msk_1.map emd_16016_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
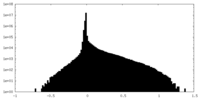
| Density Histograms |
-Additional map: Unsharpened map, output of CryoSPARC v3.4.0 local refinement job
| File | emd_16016_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map, output of CryoSPARC v3.4.0 local refinement job | ||||||||||||
| Projections & Slices |
| ||||||||||||
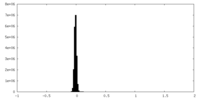
| Density Histograms |
-Half map: Half map B, output of CryoSPARC v3.4.0 local refinement job
| File | emd_16016_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B, output of CryoSPARC v3.4.0 local refinement job | ||||||||||||
| Projections & Slices |
| ||||||||||||
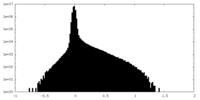
| Density Histograms |
-Half map: Half map A, output of CryoSPARC v3.4.0 local refinement job
| File | emd_16016_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A, output of CryoSPARC v3.4.0 local refinement job | ||||||||||||
| Projections & Slices |
| ||||||||||||
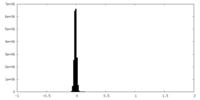
| Density Histograms |
- Sample components
Sample components
-Entire : ISDra2 TnpB binary complex
| Entire | Name: ISDra2 TnpB binary complex |
|---|---|
| Components |
|
-Supramolecule #1: ISDra2 TnpB binary complex
| Supramolecule | Name: ISDra2 TnpB binary complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: ISDra2 TnpB in complex with reRNA |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans (radioresistant) Deinococcus radiodurans (radioresistant) |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: RNA-guided DNA endonuclease TnpB
| Macromolecule | Name: RNA-guided DNA endonuclease TnpB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 (radioresistant) Deinococcus radiodurans R1 (radioresistant)Strain: ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422 |
| Molecular weight | Theoretical: 46.484289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIRNKAFVVR LYPNAAQTEL INRTLGSARF VYNHFLARRI AAYKESGKGL TYGQTSSELT LLKQAEETSW LSEVDKFALQ NSLKNLETA YKNFFRTVKQ SGKKVGFPRF RKKRTGESYR TQFTNNNIQI GEGRLKLPKL GWVKTKGQQD IQGKILNVTV R RIHEGHYE ...String: MIRNKAFVVR LYPNAAQTEL INRTLGSARF VYNHFLARRI AAYKESGKGL TYGQTSSELT LLKQAEETSW LSEVDKFALQ NSLKNLETA YKNFFRTVKQ SGKKVGFPRF RKKRTGESYR TQFTNNNIQI GEGRLKLPKL GWVKTKGQQD IQGKILNVTV R RIHEGHYE ASVLCEVEIP YLPAAPKFAA GVDVGIKDFA IVTDGVRFKH EQNPKYYRST LKRLRKAQQT LSRRKKGSAR YG KAKTKLA RIHKRIVNKR QDFLHKLTTS LVREYEIIGT EHLKPDNMRK NRRLALSISD AGWGEFIRQL EYKAAWYGRL VSK VSPYFP SSQLCHDCGF KNPEVKNLAV RTWTCPNCGE THDRDENAAL NIRREALVAA GISDTLNAHG GYVRPASAGN GLRS ENHAT LVV UniProtKB: RNA-guided DNA endonuclease TnpB |
-Macromolecule #2: Deinococcus radiodurans R1 chromosome 1
| Macromolecule | Name: Deinococcus radiodurans R1 chromosome 1 / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 (radioresistant) Deinococcus radiodurans R1 (radioresistant) |
| Molecular weight | Theoretical: 48.408633 KDa |
| Sequence | String: CAUUCGGCGU GAAGCGUUGG UGGCUGCGGG AAUCUCAGAC ACCUUAAACG CUCAUGGAGG CUAUGUCAGA CCUGCUUCGG CGGGCAAUG GUCUGCGAAG UGAGAAUCAC GCGACUUUAG UCGUGUGAGG UUCAAGAGUC CCUUGGCGCC C GENBANK: GENBANK: AE000513.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Number real images: 6688 / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)