[English] 日本語
 Yorodumi
Yorodumi- EMDB-1593: Electron crystallographic reconstruction of human copper transpor... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1593 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron crystallographic reconstruction of human copper transporter (hCTR1). | |||||||||
 Map data Map data | Volume map of human copper transporter (hCTR1) metal free. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / electron crystallography / 2D-crystal / membrane protein / copper | |||||||||
| Function / homology |  Function and homology information Function and homology informationsilver ion transmembrane transporter activity / plasma membrane copper ion transport / silver ion transmembrane transport / Metal ion SLC transporters / copper ion transmembrane transporter activity / copper ion import / vascular endothelial growth factor receptor-2 signaling pathway / copper ion transport / xenobiotic transport / protein complex oligomerization ...silver ion transmembrane transporter activity / plasma membrane copper ion transport / silver ion transmembrane transport / Metal ion SLC transporters / copper ion transmembrane transporter activity / copper ion import / vascular endothelial growth factor receptor-2 signaling pathway / copper ion transport / xenobiotic transport / protein complex oligomerization / intercalated disc / intracellular copper ion homeostasis / xenobiotic transmembrane transporter activity / establishment of localization in cell / recycling endosome membrane / late endosome membrane / early endosome membrane / angiogenesis / basolateral plasma membrane / apical plasma membrane / copper ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron crystallography / cryo EM / negative staining / Resolution: 7.0 Å | |||||||||
 Authors Authors | DeFeo CJ / Aller SG / Siluvai GS / Blackburn NJ / Unger VM | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2009 Journal: Proc Natl Acad Sci U S A / Year: 2009Title: Three-dimensional structure of the human copper transporter hCTR1. Authors: Christopher J De Feo / Stephen G Aller / Gnana S Siluvai / Ninian J Blackburn / Vinzenz M Unger /  Abstract: Copper uptake proteins (CTRs), mediate cellular acquisition of the essential metal copper in all eukaryotes. Here, we report the structure of the human CTR1 protein solved by electron crystallography ...Copper uptake proteins (CTRs), mediate cellular acquisition of the essential metal copper in all eukaryotes. Here, we report the structure of the human CTR1 protein solved by electron crystallography to an in plane resolution of 7 A. Reminiscent of the design of traditional ion channels, trimeric hCTR1 creates a pore that stretches across the membrane bilayer at the interface between the subunits. Assignment of the helices identifies the second transmembrane helix as the key element lining the pore, and reveals how functionally important residues on this helix could participate in Cu(I)-coordination during transport. Aligned with and sealing both ends of the pore, extracellular and intracellular domains of hCTR1 appear to provide additional metal binding sites. Consistent with the existence of distinct metal binding sites, we demonstrate that hCTR1 stably binds 2 Cu(I)-ions through 3-coordinate Cu-S bonds, and that mutations in one of these putative binding sites results in a change of coordination chemistry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1593.map.gz emd_1593.map.gz | 14.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1593-v30.xml emd-1593-v30.xml emd-1593.xml emd-1593.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1593.gif 1593.gif | 36.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1593 http://ftp.pdbj.org/pub/emdb/structures/EMD-1593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1593 | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1593.map.gz / Format: CCP4 / Size: 22.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1593.map.gz / Format: CCP4 / Size: 22.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Volume map of human copper transporter (hCTR1) metal free. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 1.01136 Å / Y: 1.01136 Å / Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
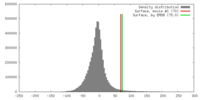
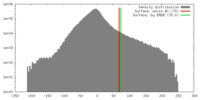
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 177 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human copper transporter hCTR1
| Entire | Name: human copper transporter hCTR1 |
|---|---|
| Components |
|
-Supramolecule #1000: human copper transporter hCTR1
| Supramolecule | Name: human copper transporter hCTR1 / type: sample / ID: 1000 / Oligomeric state: trimer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 66 KDa |
-Macromolecule #1: hCTR1
| Macromolecule | Name: hCTR1 / type: protein_or_peptide / ID: 1 / Name.synonym: copper transporter / Number of copies: 3 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: Plasma Membrane Homo sapiens (human) / synonym: Human / Location in cell: Plasma Membrane |
| Molecular weight | Theoretical: 66 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) / Recombinant plasmid: pPic3.5 Komagataella pastoris (fungus) / Recombinant plasmid: pPic3.5 |
| Sequence | UniProtKB: High affinity copper uptake protein 1 |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 0.37 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 10mM MOPS, 280mM NaCl, 2.0 mM EDTA |
| Staining | Type: NEGATIVE / Details: none |
| Grid | Details: 300 mesh molybdenum grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 93 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Home built plunger. no special treatment Method: Blot for 5 seconds before plunging |
| Details | grown by dialysis |
| Crystal formation | Details: grown by dialysis |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 93 K / Max: 93 K / Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: 230,000 times magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7.0 µm / Number real images: 58 / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan Side Entry / Specimen holder model: GATAN LIQUID NITROGEN / Tilt angle min: 1.2 / Tilt angle max: 46.2 / Tilt series - Axis1 - Min angle: 1.2 ° / Tilt series - Axis1 - Max angle: 46.2 ° |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.0 Å / Resolution method: OTHER / Software - Name: MRC |
|---|---|
| Crystal parameters | Unit cell - A: 89 Å / Unit cell - B: 89 Å / Unit cell - C: 400 Å / Unit cell - γ: 120 ° / Unit cell - α: 90 ° / Unit cell - β: 90 ° / Plane group: P 6 2 2 |
| CTF correction | Details: Each image |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)