[English] 日本語
 Yorodumi
Yorodumi- EMDB-15858: Negative stain EM map of human RE1-silencing transcription factor -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM map of human RE1-silencing transcription factor | ||||||||||||
 Map data Map data | REST negative staining EM map | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
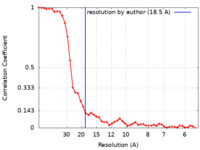
| Method | single particle reconstruction / negative staining / Resolution: 18.5 Å | ||||||||||||
 Authors Authors | Pinkas M / Brom T / Novacek J / Veverka P / Janovic T / Stojaspal M / Hofr C | ||||||||||||
| Funding support |  Czech Republic, 3 items Czech Republic, 3 items
| ||||||||||||
 Citation Citation |  Journal: Comput Struct Biotechnol J / Year: 2023 Journal: Comput Struct Biotechnol J / Year: 2023Title: Electron microscopy reveals toroidal shape of master neuronal cell differentiator REST - RE1-silencing transcription factor. Authors: Pavel Veverka / Tomáš Brom / Tomáš Janovič / Martin Stojaspal / Matyáš Pinkas / Jiří Nováček / Ctirad Hofr /  Abstract: The RE1-Silencing Transcription factor (REST) is essential for neuronal differentiation. Here, we report the first 18.5-angstrom electron microscopy structure of human REST. The refined electron map ...The RE1-Silencing Transcription factor (REST) is essential for neuronal differentiation. Here, we report the first 18.5-angstrom electron microscopy structure of human REST. The refined electron map suggests that REST forms a torus that can accommodate DNA double-helix in the central hole. Additionally, we quantitatively described REST binding to the canonical DNA sequence of the neuron-restrictive silencer element. We developed protocols for the expression and purification of full-length REST and the shortened variant REST-N62 produced by alternative splicing. We tested the mutual interaction of full-length REST and the splicing variant REST-N62. Revealed structure-function relationships of master neuronal repressor REST will allow finding new biological ways of prevention and treatment of neurodegenerative disorders and diseases. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15858.map.gz emd_15858.map.gz | 6.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15858-v30.xml emd-15858-v30.xml emd-15858.xml emd-15858.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15858_fsc.xml emd_15858_fsc.xml | 4.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15858.png emd_15858.png | 69.4 KB | ||
| Others |  emd_15858_half_map_1.map.gz emd_15858_half_map_1.map.gz emd_15858_half_map_2.map.gz emd_15858_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15858 http://ftp.pdbj.org/pub/emdb/structures/EMD-15858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15858 | HTTPS FTP |
-Validation report
| Summary document |  emd_15858_validation.pdf.gz emd_15858_validation.pdf.gz | 496.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15858_full_validation.pdf.gz emd_15858_full_validation.pdf.gz | 495.7 KB | Display | |
| Data in XML |  emd_15858_validation.xml.gz emd_15858_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_15858_validation.cif.gz emd_15858_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15858 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15858 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15858.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15858.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | REST negative staining EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.647 Å | ||||||||||||||||||||||||||||||||||||
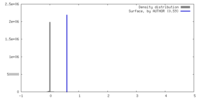
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: REST half1 map
| File | emd_15858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | REST half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
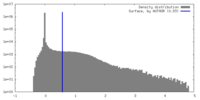
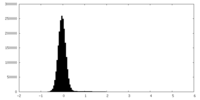
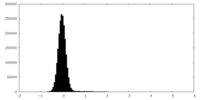
| Density Histograms |
-Half map: REST half2 map
| File | emd_15858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | REST half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
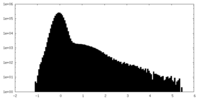
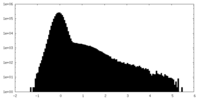
| Density Histograms |
- Sample components
Sample components
-Entire : RE1-silencing transcription factor
| Entire | Name: RE1-silencing transcription factor |
|---|---|
| Components |
|
-Supramolecule #1: RE1-silencing transcription factor
| Supramolecule | Name: RE1-silencing transcription factor / type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Location in cell: nucleus, cytoplasm Homo sapiens (human) / Location in cell: nucleus, cytoplasm |
| Molecular weight | Theoretical: 122 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Buffer was prepared fresh then it was filtered by 0,22 um filter and degassed using ultrasound. | |||||||||
| Staining | Type: NEGATIVE / Material: uranyl acetate | |||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 12.0 nm / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS F200C |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Average electron dose: 15.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)