[English] 日本語
 Yorodumi
Yorodumi- EMDB-15831: CryoEM structure of the pointy tip (proteins pIII/pVI/pVIII) from... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the pointy tip (proteins pIII/pVI/pVIII) from the f1 filamentous bacteriophage | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viral proteins / Infection / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral extrusion / virion attachment to host cell pilus / adhesion receptor-mediated virion attachment to host cell / helical viral capsid / host cell membrane / virion component / viral capsid / entry receptor-mediated virion attachment to host cell / symbiont entry into host cell / membrane Similarity search - Function | |||||||||
| Biological species |  Enterobacteria phage f1 (virus) Enterobacteria phage f1 (virus) | |||||||||
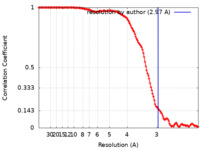
| Method | single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | Conners R / McLaren M / Gold VAM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-electron microscopy of the f1 filamentous phage reveals insights into viral infection and assembly. Authors: Rebecca Conners / Rayén Ignacia León-Quezada / Mathew McLaren / Nicholas J Bennett / Bertram Daum / Jasna Rakonjac / Vicki A M Gold /   Abstract: Phages are viruses that infect bacteria and dominate every ecosystem on our planet. As well as impacting microbial ecology, physiology and evolution, phages are exploited as tools in molecular ...Phages are viruses that infect bacteria and dominate every ecosystem on our planet. As well as impacting microbial ecology, physiology and evolution, phages are exploited as tools in molecular biology and biotechnology. This is particularly true for the Ff (f1, fd or M13) phages, which represent a widely distributed group of filamentous viruses. Over nearly five decades, Ffs have seen an extraordinary range of applications, yet the complete structure of the phage capsid and consequently the mechanisms of infection and assembly remain largely mysterious. In this work, we use cryo-electron microscopy and a highly efficient system for production of short Ff-derived nanorods to determine a structure of a filamentous virus including the tips. We show that structure combined with mutagenesis can identify phage domains that are important in bacterial attack and for release of new progeny, allowing new models to be proposed for the phage lifecycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15831.map.gz emd_15831.map.gz | 108.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15831-v30.xml emd-15831-v30.xml emd-15831.xml emd-15831.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15831_fsc.xml emd_15831_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_15831.png emd_15831.png | 45.2 KB | ||
| Filedesc metadata |  emd-15831.cif.gz emd-15831.cif.gz | 5.9 KB | ||
| Others |  emd_15831_half_map_1.map.gz emd_15831_half_map_1.map.gz emd_15831_half_map_2.map.gz emd_15831_half_map_2.map.gz | 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15831 http://ftp.pdbj.org/pub/emdb/structures/EMD-15831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15831 | HTTPS FTP |
-Validation report
| Summary document |  emd_15831_validation.pdf.gz emd_15831_validation.pdf.gz | 824.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15831_full_validation.pdf.gz emd_15831_full_validation.pdf.gz | 823.7 KB | Display | |
| Data in XML |  emd_15831_validation.xml.gz emd_15831_validation.xml.gz | 21.5 KB | Display | |
| Data in CIF |  emd_15831_validation.cif.gz emd_15831_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15831 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15831 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15831 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15831 | HTTPS FTP |
-Related structure data
| Related structure data |  8b3oMC  8b3pC  8b3qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15831.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15831.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1012 Å | ||||||||||||||||||||||||||||||||||||
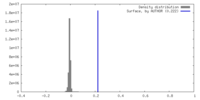
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15831_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
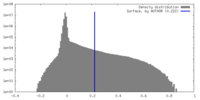
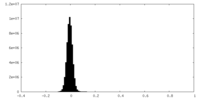
| Density Histograms |
-Half map: #2
| File | emd_15831_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
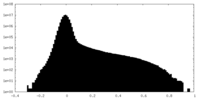
| Density Histograms |
- Sample components
Sample components
-Entire : Enterobacteria phage f1
| Entire | Name:  Enterobacteria phage f1 (virus) Enterobacteria phage f1 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage f1
| Supramolecule | Name: Enterobacteria phage f1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10863 / Sci species name: Enterobacteria phage f1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: Capsid protein G8P
| Macromolecule | Name: Capsid protein G8P / type: protein_or_peptide / ID: 1 / Number of copies: 35 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage f1 (virus) Enterobacteria phage f1 (virus) |
| Molecular weight | Theoretical: 5.212021 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AEGDDPAKAA FDSLQASATE MIGYAWAMVV VIVGATIGIK LFKKFTSKAS UniProtKB: Capsid protein G8P |
-Macromolecule #2: Head virion protein G6P
| Macromolecule | Name: Head virion protein G6P / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage f1 (virus) Enterobacteria phage f1 (virus) |
| Molecular weight | Theoretical: 12.357984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPVLLGIPLL LRFLGFLLVT LFGYLLTFLK KGFGKIAIAI SLFLALIIGL NSILVGYLSD ISAQLPSDFV QGVQLILPSN ALPCFYVIL SVKAAIFIFD VKQKIVSYLD WDK UniProtKB: Head virion protein G6P |
-Macromolecule #3: Attachment protein G3P
| Macromolecule | Name: Attachment protein G3P / type: protein_or_peptide / ID: 3 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage f1 (virus) Enterobacteria phage f1 (virus) |
| Molecular weight | Theoretical: 42.573766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AETVESCLAK PHTENSFTNV WKDDKTLDRY ANYEGCLWNA TGVVVCTGDE TQCYGTWVPI GLAIPENEGG GSEGGGSEGG GSEGGGTKP PEYGDTPIPG YTYINPLDGT YPPGTEQNPA NPNPSLEESQ PLNTFMFQNN RFRNRQGALT VYTGTVTQGT D PVKTYYQY ...String: AETVESCLAK PHTENSFTNV WKDDKTLDRY ANYEGCLWNA TGVVVCTGDE TQCYGTWVPI GLAIPENEGG GSEGGGSEGG GSEGGGTKP PEYGDTPIPG YTYINPLDGT YPPGTEQNPA NPNPSLEESQ PLNTFMFQNN RFRNRQGALT VYTGTVTQGT D PVKTYYQY TPVSSKAMYD AYWNGKFRDC AFHSGFNEDP FVCEYQGQSS DLPQPPVNAG GGSGGGSGGG SEGGGSEGGG SE GGGSEGG GSGGGSGSGD FDYEKMANAN KGAMTENADE NALQSDAKGK LDSVATDYGA AIDGFIGDVS GLANGNGATG DFA GSNSQM AQVGDGDNSP LMNNFRQYLP SLPQSVECRP FVFGAGKPYE FSIDCDKINL FRGVFAFLLY VATFMYVFST FANI LRNKE S UniProtKB: Attachment protein G3P |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Wait time 5 sec., drain time 0 sec., blot force 0, blot time 4 sec.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-8b3o: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)