+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | catalytic amyloid fibril formed by Ac-LHLHLRL-amide | |||||||||
 Map data Map data | electron density map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | catalytic amyloid fibril / protein fibril / zinc binding / prion | |||||||||
| Biological species | synthetic construct (others) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
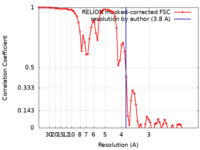
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Heerde T / Schmidt M / Faendrich M | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2023 Journal: Sci Rep / Year: 2023Title: Cryo-EM structure of a catalytic amyloid fibril. Authors: Thomas Heerde / Akanksha Bansal / Matthias Schmidt / Marcus Fändrich /  Abstract: Catalytic amyloid fibrils are novel types of bioinspired, functional materials that combine the chemical and mechanical robustness of amyloids with the ability to catalyze a certain chemical reaction. ...Catalytic amyloid fibrils are novel types of bioinspired, functional materials that combine the chemical and mechanical robustness of amyloids with the ability to catalyze a certain chemical reaction. In this study we used cryo-electron microcopy to analyze the amyloid fibril structure and the catalytic center of amyloid fibrils that hydrolyze ester bonds. Our findings show that catalytic amyloid fibrils are polymorphic and consist of similarly structured, zipper-like building blocks that consist of mated cross-β sheets. These building blocks define the fibril core, which is decorated by a peripheral leaflet of peptide molecules. The observed structural arrangement differs from previously described catalytic amyloid fibrils and yielded a new model of the catalytic center. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15824.map.gz emd_15824.map.gz | 694.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15824-v30.xml emd-15824-v30.xml emd-15824.xml emd-15824.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15824_fsc.xml emd_15824_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15824.png emd_15824.png | 34 KB | ||
| Masks |  emd_15824_msk_1.map emd_15824_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15824.cif.gz emd-15824.cif.gz | 4.9 KB | ||
| Others |  emd_15824_half_map_1.map.gz emd_15824_half_map_1.map.gz emd_15824_half_map_2.map.gz emd_15824_half_map_2.map.gz | 22.5 MB 22.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15824 http://ftp.pdbj.org/pub/emdb/structures/EMD-15824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15824 | HTTPS FTP |
-Validation report
| Summary document |  emd_15824_validation.pdf.gz emd_15824_validation.pdf.gz | 623.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15824_full_validation.pdf.gz emd_15824_full_validation.pdf.gz | 623.2 KB | Display | |
| Data in XML |  emd_15824_validation.xml.gz emd_15824_validation.xml.gz | 12.8 KB | Display | |
| Data in CIF |  emd_15824_validation.cif.gz emd_15824_validation.cif.gz | 17.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15824 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15824 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15824 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15824 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15824.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15824.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | electron density map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15824_msk_1.map emd_15824_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
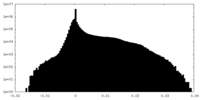
| Density Histograms |
-Half map: first half-map of the 3D refinement
| File | emd_15824_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | first half-map of the 3D refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
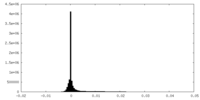
| Density Histograms |
-Half map: second half-map of the 3D refinement
| File | emd_15824_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | second half-map of the 3D refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
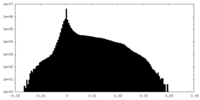
| Density Histograms |
- Sample components
Sample components
-Entire : catalytic amyloid
| Entire | Name: catalytic amyloid |
|---|---|
| Components |
|
-Supramolecule #1: catalytic amyloid
| Supramolecule | Name: catalytic amyloid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: ACE-LEU-HIS-LEU-HIS-LEU-ARG-LEU-NH2
| Macromolecule | Name: ACE-LEU-HIS-LEU-HIS-LEU-ARG-LEU-NH2 / type: protein_or_peptide / ID: 1 / Number of copies: 30 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 928.179 Da |
| Sequence | String: (ACE)LHLHLRL(NH2) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.0471 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 25 mM Tris(hydroxymethyl)aminomethane (Tris), 1 mM Zincchloride | |||||||||
| Grid | Model: C-flat-1.2/1.3 / Mesh: 400 | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 12.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Target criteria: correlation coefficient |
|---|---|
| Output model |  PDB-8b3a: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)