+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human adenovirus type 5 lacking core protein V | |||||||||
 Map data Map data | Human adenovirus type 5 lacking core protein V | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HAdV-C5 / deltaV / VIRUS | |||||||||
| Biological species |  Ad5 deltaV (virus) Ad5 deltaV (virus) | |||||||||
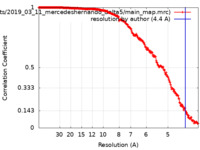
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Hernando-Perez M / San Martin C / Martin-Gonzalez N / Gomez-Gonzalez A / Bauer M / Greber UF / de Pablo PJ | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Adenovirus core protein V reinforces the capsid and enhances genome release from disrupted particles. Authors: Natalia Martín-González / Alfonso Gómez-González / Mercedes Hernando-Pérez / Michael Bauer / Urs F Greber / Carmen San Martín / Pedro J de Pablo /   Abstract: Out of the three core proteins in human adenovirus, protein V is believed to connect the inner capsid surface to the outer genome layer. Here, we explored mechanical properties and in vitro ...Out of the three core proteins in human adenovirus, protein V is believed to connect the inner capsid surface to the outer genome layer. Here, we explored mechanical properties and in vitro disassembly of particles lacking protein V (Ad5-ΔV). Ad5-ΔV particles were softer and less brittle than the wild-type ones (Ad5-wt), but they were more prone to release pentons under mechanical fatigue. In Ad5-ΔV, core components did not readily diffuse out of partially disrupted capsids, and the core appeared more condensed than in Ad5-wt. These observations suggest that instead of condensing the genome, protein V antagonizes the condensing action of the other core proteins. Protein V provides mechanical reinforcement and facilitates genome release by keeping DNA connected to capsid fragments that detach during disruption. This scenario is in line with the location of protein V in the virion and its role in Ad5 cell entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15694.map.gz emd_15694.map.gz | 882.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15694-v30.xml emd-15694-v30.xml emd-15694.xml emd-15694.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15694_fsc.xml emd_15694_fsc.xml | 18.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15694.png emd_15694.png | 243.8 KB | ||
| Others |  emd_15694_half_map_1.map.gz emd_15694_half_map_1.map.gz emd_15694_half_map_2.map.gz emd_15694_half_map_2.map.gz | 801.5 MB 801.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15694 http://ftp.pdbj.org/pub/emdb/structures/EMD-15694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15694 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15694.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15694.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human adenovirus type 5 lacking core protein V | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.025 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half2 map in the same coordinate space as the primary map
| File | emd_15694_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 map in the same coordinate space as the primary map | ||||||||||||
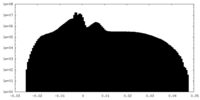
| Projections & Slices |
| ||||||||||||
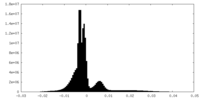
| Density Histograms |
-Half map: Half1 map in the same coordinate space as the primary map
| File | emd_15694_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 map in the same coordinate space as the primary map | ||||||||||||
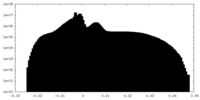
| Projections & Slices |
| ||||||||||||
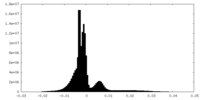
| Density Histograms |
- Sample components
Sample components
-Entire : Ad5 deltaV
| Entire | Name:  Ad5 deltaV (virus) Ad5 deltaV (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Ad5 deltaV
| Supramolecule | Name: Ad5 deltaV / type: virus / ID: 1 / Parent: 0 Details: Deletion mutant described in https://doi.org/10.1126/sciadv.abl7150 NCBI-ID: 129951 / Sci species name: Ad5 deltaV / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 MDa |
| Virus shell | Shell ID: 1 / Diameter: 950.0 Å / T number (triangulation number): 25 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 2106 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.442 µm / Calibrated defocus min: 0.483 µm / Calibrated magnification: 73000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 73000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)