[English] 日本語
 Yorodumi
Yorodumi- EMDB-15654: Structure of the giant inhibitor of apoptosis, BIRC6 bound to the... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the giant inhibitor of apoptosis, BIRC6 bound to the regulator SMAC | |||||||||
 Map data Map data | Sharpened map from non-uniform refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E2/E3 ubiquitin ligase / APOPTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationRelease of apoptotic factors from the mitochondria / CD40 receptor complex / labyrinthine layer development / (E3-independent) E2 ubiquitin-conjugating enzyme / ALK mutants bind TKIs / SMAC, XIAP-regulated apoptotic response / Flemming body / Regulation of the apoptosome activity / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes ...Release of apoptotic factors from the mitochondria / CD40 receptor complex / labyrinthine layer development / (E3-independent) E2 ubiquitin-conjugating enzyme / ALK mutants bind TKIs / SMAC, XIAP-regulated apoptotic response / Flemming body / Regulation of the apoptosome activity / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / intrinsic apoptotic signaling pathway in response to oxidative stress / microtubule organizing center / ubiquitin conjugating enzyme activity / extrinsic apoptotic signaling pathway via death domain receptors / cysteine-type endopeptidase inhibitor activity / intrinsic apoptotic signaling pathway / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / trans-Golgi network / mitochondrial intermembrane space / cytoplasmic side of plasma membrane / spindle pole / ubiquitin-protein transferase activity / Signaling by ALK fusions and activated point mutants / regulation of cell population proliferation / midbody / neuron apoptotic process / cell population proliferation / endosome / protein ubiquitination / protein phosphorylation / positive regulation of apoptotic process / cell division / apoptotic process / positive regulation of cell population proliferation / centrosome / negative regulation of apoptotic process / mitochondrion / metal ion binding / nucleus / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Dietz L / Elliott PR | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural basis for SMAC-mediated antagonism of caspase inhibition by the giant ubiquitin ligase BIRC6. Authors: Larissa Dietz / Cara J Ellison / Carlos Riechmann / C Keith Cassidy / F Daniel Felfoldi / Adán Pinto-Fernández / Benedikt M Kessler / Paul R Elliott /  Abstract: Certain inhibitor of apoptosis (IAP) family members are sentinel proteins that prevent untimely cell death by inhibiting caspases. Antagonists, including second mitochondria-derived activator of ...Certain inhibitor of apoptosis (IAP) family members are sentinel proteins that prevent untimely cell death by inhibiting caspases. Antagonists, including second mitochondria-derived activator of caspases (SMAC), regulate IAPs and drive cell death. Baculoviral IAP repeat-containing protein 6 (BIRC6), a giant IAP with dual E2 and E3 ubiquitin ligase activity, regulates programmed cell death through unknown mechanisms. We show that BIRC6 directly restricts executioner caspase-3 and -7 and ubiquitinates caspase-3, -7, and -9, working exclusively with noncanonical E1, UBA6. Notably, we show that SMAC suppresses both mechanisms. Cryo-electron microscopy structures of BIRC6 alone and in complex with SMAC reveal that BIRC6 is an antiparallel dimer juxtaposing the substrate-binding module against the catalytic domain. Furthermore, we discover that SMAC multisite binding to BIRC6 results in a subnanomolar affinity interaction, enabling SMAC to competitively displace caspases, thus antagonizing BIRC6 anticaspase function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15654.map.gz emd_15654.map.gz | 95.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15654-v30.xml emd-15654-v30.xml emd-15654.xml emd-15654.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15654.png emd_15654.png | 74.8 KB | ||
| Masks |  emd_15654_msk_1.map emd_15654_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15654.cif.gz emd-15654.cif.gz | 9 KB | ||
| Others |  emd_15654_half_map_1.map.gz emd_15654_half_map_1.map.gz emd_15654_half_map_2.map.gz emd_15654_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15654 http://ftp.pdbj.org/pub/emdb/structures/EMD-15654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15654 | HTTPS FTP |
-Related structure data
| Related structure data |  8atoMC  8atmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15654.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15654.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from non-uniform refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.829 Å | ||||||||||||||||||||||||||||||||||||
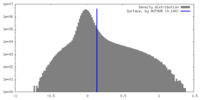
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15654_msk_1.map emd_15654_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
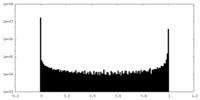
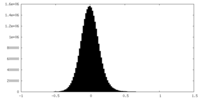
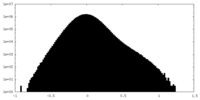
| Density Histograms |
-Half map: First half map non-uniform refinement
| File | emd_15654_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
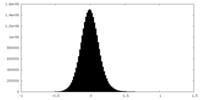
| Density Histograms |
-Half map: Second half map non-uniform refinement
| File | emd_15654_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
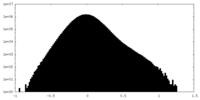
| Density Histograms |
- Sample components
Sample components
-Entire : Anti-parallel homodimer
| Entire | Name: Anti-parallel homodimer |
|---|---|
| Components |
|
-Supramolecule #1: Anti-parallel homodimer
| Supramolecule | Name: Anti-parallel homodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 1 MDa |
-Supramolecule #2: Baculoviral IAP repeat-containing protein 6
| Supramolecule | Name: Baculoviral IAP repeat-containing protein 6 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Diablo IAP-binding mitochondrial protein
| Supramolecule | Name: Diablo IAP-binding mitochondrial protein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Baculoviral IAP repeat-containing protein 6
| Macromolecule | Name: Baculoviral IAP repeat-containing protein 6 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 530.977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPMVTGGGAA PPGTVTEPLP SVIVLSAGRK MAAAAAAASG PGCSSAAGAG AAGVSEWLVL RDGCMHCDAD GLHSLSYHPA LNAILAVTS RGTIKVIDGT SGATLQASAL SAKPGGQVKC QYISAVDKVI FVDDYAVGCR KDLNGILLLD TALQTPVSKQ D DVVQLELP ...String: GPMVTGGGAA PPGTVTEPLP SVIVLSAGRK MAAAAAAASG PGCSSAAGAG AAGVSEWLVL RDGCMHCDAD GLHSLSYHPA LNAILAVTS RGTIKVIDGT SGATLQASAL SAKPGGQVKC QYISAVDKVI FVDDYAVGCR KDLNGILLLD TALQTPVSKQ D DVVQLELP VTEAQQLLSA CLEKVDISST EGYDLFITQL KDGLKNTSHE TAANHKVAKW ATVTFHLPHH VLKSIASAIV NE LKKINQN VAALPVASSV MDRLSYLLPS ARPELGVGPG RSVDRSLMYS EANRRETFTS WPHVGYRWAQ PDPMAQAGFY HQP ASSGDD RAMCFTCSVC LVCWEPTDEP WSEHERHSPN CPFVKGEHTQ NVPLSVTLAT SPAQFPCTDG TDRISCFGSG SCPH FLAAA TKRGKICIWD VSKLMKVHLK FEINAYDPAI VQQLILSGDP SSGVDSRRPT LAWLEDSSSC SDIPKLEGDS DDLLE DSDS EEHSRSDSVT GHTSQKEAME VSLDITALSI LQQPEKLQWE IVANVLEDTV KDLEELGANP CLTNSKSEKT KEKHQE QHN IPFPCLLAGG LLTYKSPATS PISSNSHRSL DGLSRTQGES ISEQGSTDNE SCTNSELNSP LVRRTLPVLL LYSIKES DE KAGKIFSQMN NIMSKSLHDD GFTVPQIIEM ELDSQEQLLL QDPPVTYIQQ FADAAANLTS PDSEKWNSVF PKPGTLVQ C LRLPKFAEEE NLCIDSITPC ADGIHLLVGL RTCPVESLSA INQVEALNNL NKLNSALCNR RKGELESNLA VVNGANISV IQHESPADVQ TPLIIQPEQR NVSGGYLVLY KMNYATRIVT LEEEPIKIQH IKDPQDTITS LILLPPDILD NREDDCEEPI EDMQLTSKN GFEREKTSDI STLGHLVITT QGGYVKILDL SNFEILAKVE PPKKEGTEEQ DTFVSVIYCS GTDRLCACTK G GELHFLQI GGTCDDIDEA DILVDGSLSK GIEPSSEGSK PLSNPSSPGI SGVDLLVDQP FTLEILTSLV ELTRFETLTP RF SATVPPC WVEVQQEQQQ RRHPQHLHQQ HHGDAAQHTR TWKLQTDSNS WDEHVFELVL PKACMVGHVD FKFVLNSNIT NIP QIQVTL LKNKAPGLGK VNALNIEVEQ NGKPSLVDLN EEMQHMDVEE SQCLRLCPFL EDHKEDILCG PVWLASGLDL SGHA GMLTL TSPKLVKGMA GGKYRSFLIH VKAVNERGTE EICNGGMRPV VRLPSLKHQS NKGYSLASLL AKVAAGKEKS SNVKN ENTS GTRKSENLRG CDLLQEVSVT IRRFKKTSIS KERVQRCAML QFSEFHEKLL NTLCRKTDDG QITEHAQSLV LDTLCW LAG VHSNGPGSSK EGNENLLSKT RKFLSDIVRV CFFEAGRSIA HKCARFLALC ISNGKCDPCQ PAFGPVLLKA LLDNMSF LP AATTGGSVYW YFVLLNYVKD EDLAGCSTAC ASLLTAVSRQ LQDRLTPMEA LLQTRYGLYS SPFDPVLFDL EMSGSSCK N VYNSSIGVQS DEIDLSDVLS GNGKVSSCTA AEGSFTSLTG LLEVEPLHFT CVSTSDGTRI ERDDAMSSFG VTPAVGGLS SGTVGEASTA LSSAAQVALQ SLSHAMASAE QQLQVLQEKQ QQLLKLQQQK AKLEAKLHQT TAAAAAAASA VGPVHNSVPS NPVAAPGFF IHPSDVIPPT PKTTPLFMTP PLTPPNEAVS VVINAELAQL FPGSVIDPPA VNLAAHNKNS NKSRMNPLGS G LALAISHA SHFLQPPPHQ SIIIERMHSG ARRFVTLDFG RPILLTDVLI PTCGDLASLS IDIWTLGEEV DGRRLVVATD IS THSLILH DLIPPPVCRF MKITVIGRYG STNARAKIPL GFYYGHTYIL PWESELKLMH DPLKGEGESA NQPEIDQHLA MMV ALQEDI QCRYNLACHR LETLLQSIDL PPLNSANNAQ YFLRKPDKAV EEDSRVFSAY QDCIQLQLQL NLAHNAVQRL KVAL GASRK MLSETSNPED LIQTSSTEQL RTIIRYLLDT LLSLLHASNG HSVPAVLQST FHAQACEELF KHLCISGTPK IRLHT GLLL VQLCGGERWW GQFLSNVLQE LYNSEQLLIF PQDRVFMLLS CIGQRSLSNS GVLESLLNLL DNLLSPLQPQ LPMHRR TEG VLDIPMISWV VMLVSRLLDY VATVEDEAAA AKKPLNGNQW SFINNNLHTQ SLNRSSKGSS SLDRLYSRKI RKQLVHH KQ QLNLLKAKQK ALVEQMEKEK IQSNKGSSYK LLVEQAKLKQ ATSKHFKDLI RLRRTAEWSR SNLDTEVTTA KESPEIEP L PFTLAHERCI SVVQKLVLFL LSMDFTCHAD LLLFVCKVLA RIANATRPTI HLCEIVNEPQ LERLLLLLVG TDFNRGDIS WGGAWAQYSL TCMLQDILAG ELLAPVAAEA MEEGTVGDDV GATAGDSDDS LQQSSVQLLE TIDEPLTHDI TGAPPLSSLE KDKEIDLEL LQDLMEVDID PLDIDLEKDP LAAKVFKPIS STWYDYWGAD YGTYNYNPYI GGLGIPVAKP PANTEKNGSQ T VSVSVSQA LDARLEVGLE QQAELMLKMM STLEADSILQ ALTNTSPTLS QSPTGTDDSL LGGLQAANQT SQLIIQLSSV PM LNVCFNK LFSMLQVHHV QLESLLQLWL TLSLNSSSTG NKENGADIFL YNANRIPVIS LNQASITSFL TVLAWYPNTL LRT WCLVLH SLTLMTNMQL NSGSSSAIGT QESTAHLLVS DPNLIHVLVK FLSGTSPHGT NQHSPQVGPT ATQAMQEFLT RLQV HLSST CPQIFSEFLL KLIHILSTER GAFQTGQGPL DAQVKLLEFT LEQNFEVVSV STISAVIESV TFLVHHYITC SDKVM SRSG SDSSVGARAC FGGLFANLIR PGDAKAVCGE MTRDQLMFDL LKLVNILVQL PLSGNREYSA RVSVTTNTTD SVSDEE KVS GGKDGNGSST SVQGSPAYVA DLVLANQQIM SQILSALGLC NSSAMAMIIG ASGLHLTKHE NFHGGLDAIS VGDGLFT IL TTLSKKASTV HMMLQPILTY MACGYMGRQG SLATCQLSEP LLWFILRVLD TSDALKAFHD MGGVQLICNN MVTSTRAI V NTARSMVSTI MKFLDSGPNK AVDSTLKTRI LASEPDNAEG IHNFAPLGTI TSSSPTAQPA EVLLQATPPH RRARSAAWS YIFLPEEAWC DLTIHLPAAV LLKEIHIQPH LASLATCPSS VSVEVSADGV NMLPLSTPVV TSGLTYIKIQ LVKAEVASAV CLRLHRPRD ASTLGLSQIK LLGLTAFGTT SSATVNNPFL PSEDQVSKTS IGWLRLLHHC LTHISDLEGM MASAAAPTAN L LQTCAALL MSPYCGMHSP NIEVVLVKIG LQSTRIGLKL IDILLRNCAA SGSDPTDLNS PLLFGRLNGL SSDSTIDILY QL GTTQDPG TKDRIQALLK WVSDSARVAA MKRSGRMNYM CPNSSTVEYG LLMPSPSHLH CVAAILWHSY ELLVEYDLPA LLD QELFEL LFNWSMSLPC NMVLKKAVDS LLCSMCHVHP NYFSLLMGWM GITPPPVQCH HRLSMTDDSK KQDLSSSLTD DSKN AQAPL ALTESHLATL ASSSQSPEAI KQLLDSGLPS LLVRSLASFC FSHISSSESI AQSIDISQDK LRRHHVPQQC NKMPI TADL VAPILRFLTE VGNSHIMKDW LGGSEVNPLW TALLFLLCHS GSTSGSHNLG AQQTSARSAS LSSAATTGLT TQQRTA IEN ATVAFFLQCI SCHPNNQKLM AQVLCELFQT SPQRGNLPTS GNISGFIRRL FLQLMLEDEK VTMFLQSPCP LYKGRIN AT SHVIQHPMYG AGHKFRTLHL PVSTTLSDVL DRVSDTPSIT AKLISEQKDD KEKKNHEEKE KVKAENGFQD NYSVVVAS G LKSQSKRAVS ATPPRPPSRR GRTIPDKIGS TSGAEAANKI ITVPVFHLFH KLLAGQPLPA EMTLAQLLTL LYDRKLPQG YRSIDLTVKL GSRVITDPSL SKTDSYKRLH PEKDHGDLLA SCPEDEALTP GDECMDGILD ESLLETCPIQ SPLQVFAGMG GLALIAERL PMLYPEVIQQ VSAPVVTSTT QEKPKDSDQF EWVTIEQSGE LVYEAPETVA AEPPPIKSAV QTMSPIPAHS L AAFGLFLR LPGYAEVLLK ERKHAQCLLR LVLGVTDDGE GSHILQSPSA NVLPTLPFHV LRSLFSTTPL TTDDGVLLRR MA LEIGALH LILVCLSALS HHSPRVPNSS VNQTEPQVSS SHNPTSTEEQ QLYWAKGTGF GTGSTASGWD VEQALTKQRL EEE HVTCLL QVLASYINPV SSAVNGEAQS SHETRGQNSN ALPSVLLELL SQSCLIPAMS SYLRNDSVLD MARHVPLYRA LLEL LRAIA SCAAMVPLLL PLSTENGEEE EEQSECQTSV GTLLAKMKTC VDTYTNRLRS KRENVKTGVK PDASDQEPEG LTLLV PDIQ KTAEIVYAAT TSLRQANQEK KLGEYSKKAA MKPKPLSVLK SLEEKYVAVM KKLQFDTFEM VSEDEDGKLG FKVNYH YMS QVKNANDANS AARARRLAQE AVTLSTSLPL SSSSSVFVRC DEERLDIMKV LITGPADTPY ANGCFEFDVY FPQDYPS SP PLVNLETTGG HSVRFNPNLY NDGKVCLSIL NTWHGRPEEK WNPQTSSFLQ VLVSVQSLIL VAEPYFNEPG YERSRGTP S GTQSSREYDG NIRQATVKWA MLEQIRNPSP CFKEVIHKHF YLKRVEIMAQ CEEWIADIQQ YSSDKRVGRT MSHHAAALK RHTAQLREEL LKLPCPEGLD PDTDDAPEVC RATTGAEETL MHDQVKPSSS KELPSDFQL UniProtKB: Dual E2 ubiquitin-conjugating enzyme/E3 ubiquitin-protein ligase BIRC6 |
-Macromolecule #2: Diablo IAP-binding mitochondrial protein
| Macromolecule | Name: Diablo IAP-binding mitochondrial protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.787098 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AVPIAQKSEP HSLSSEALMR RAVSLVTDST STFLSQTTYA LIEAITEYTK AVYTLTSLYR QYTSLLGKMN SEEEDEVWQV IIGARAEMT SKHQEYLKLE TTWMTAVGLS EMAAEAAYQT GADQASITAR NHIQLVKLQV EEVHQLSRKA ETKLAEAQIE E LRQKTQEE GEERAESEQE AYLRED UniProtKB: Diablo IAP-binding mitochondrial protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.3 sec. / Average electron dose: 47.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.75 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 191.06 |
|---|---|
| Output model |  PDB-8ato: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)