+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of the augmin TIII subcomplex | |||||||||
 Map data Map data | Cryo-EM reconstruction of the augmin TIII subcomplex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Microtubule / Branching / Nucleation / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationHAUS complex / microtubule minus-end binding / microtubule organizing center organization / mitotic spindle microtubule / centrosome cycle / microtubule organizing center / spindle assembly / spindle / mitotic spindle / microtubule ...HAUS complex / microtubule minus-end binding / microtubule organizing center organization / mitotic spindle microtubule / centrosome cycle / microtubule organizing center / spindle assembly / spindle / mitotic spindle / microtubule / cell division / centrosome / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species | ||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.7 Å | |||||||||
 Authors Authors | Zupa E / Pfeffer S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: The augmin complex architecture reveals structural insights into microtubule branching. Authors: Erik Zupa / Martin Würtz / Annett Neuner / Thomas Hoffmann / Mandy Rettel / Anna Böhler / Bram J A Vermeulen / Sebastian Eustermann / Elmar Schiebel / Stefan Pfeffer /  Abstract: In mitosis, the augmin complex binds to spindle microtubules to recruit the γ-tubulin ring complex (γ-TuRC), the principal microtubule nucleator, for the formation of branched microtubules. Our ...In mitosis, the augmin complex binds to spindle microtubules to recruit the γ-tubulin ring complex (γ-TuRC), the principal microtubule nucleator, for the formation of branched microtubules. Our understanding of augmin-mediated microtubule branching is hampered by the lack of structural information on the augmin complex. Here, we elucidate the molecular architecture and conformational plasticity of the augmin complex using an integrative structural biology approach. The elongated structure of the augmin complex is characterised by extensive coiled-coil segments and comprises two structural elements with distinct but complementary functions in γ-TuRC and microtubule binding, linked by a flexible hinge. The augmin complex is recruited to microtubules via a composite microtubule binding site comprising a positively charged unordered extension and two calponin homology domains. Our study provides the structural basis for augmin function in branched microtubule formation, decisively fostering our understanding of spindle formation in mitosis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15631.map.gz emd_15631.map.gz | 51.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15631-v30.xml emd-15631-v30.xml emd-15631.xml emd-15631.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15631.png emd_15631.png | 26.8 KB | ||
| Filedesc metadata |  emd-15631.cif.gz emd-15631.cif.gz | 8.2 KB | ||
| Others |  emd_15631_half_map_1.map.gz emd_15631_half_map_1.map.gz emd_15631_half_map_2.map.gz emd_15631_half_map_2.map.gz | 50.5 MB 50.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15631 http://ftp.pdbj.org/pub/emdb/structures/EMD-15631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15631 | HTTPS FTP |
-Related structure data
| Related structure data |  8at2MC  8at3C  8at4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15631.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15631.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the augmin TIII subcomplex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.14 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM reconstruction of the augmin TIII subcomplex-half map A
| File | emd_15631_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the augmin TIII subcomplex-half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
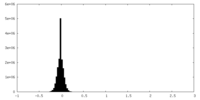
| Density Histograms |
-Half map: Cryo-EM reconstruction of the augmin TIII subcomplex-half map B
| File | emd_15631_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the augmin TIII subcomplex-half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
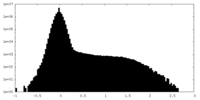
| Density Histograms |
- Sample components
Sample components
-Entire : Augmin TIII subcomplex
| Entire | Name: Augmin TIII subcomplex |
|---|---|
| Components |
|
-Supramolecule #1: Augmin TIII subcomplex
| Supramolecule | Name: Augmin TIII subcomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 218 KDa |
-Macromolecule #1: HAUS augmin-like complex subunit 1
| Macromolecule | Name: HAUS augmin-like complex subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 32.684684 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MDEKSTKIIM WLKKMFGDKP LPPYEVNTRT MEILYQLAEW NEARDKDLSL VTEDLKLKSA EVKAEAKYLQ DLLTEGLGPS YTNLSRMGN NYLNQIVDSC LALELKNSSL SSYIPAVNDL SSELVAIELN NQEMEAELTS LRKKLTEALV LEKSLERDLK K AEEQCNFE ...String: MDEKSTKIIM WLKKMFGDKP LPPYEVNTRT MEILYQLAEW NEARDKDLSL VTEDLKLKSA EVKAEAKYLQ DLLTEGLGPS YTNLSRMGN NYLNQIVDSC LALELKNSSL SSYIPAVNDL SSELVAIELN NQEMEAELTS LRKKLTEALV LEKSLERDLK K AEEQCNFE KAKVEIRSQN MKKLKDKSEE YKYKIHAAKD QLSSAGMEEP LTHRSLVSLS ETLTELKAQS MAAKEKLNSY LD LAPNPSL VKVKIEEAKR ELKATEVELT TKVNMMEFVV PEPSKRRLK UniProtKB: LOC495502 protein |
-Macromolecule #2: HAUS augmin-like complex subunit 3
| Macromolecule | Name: HAUS augmin-like complex subunit 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 68.11225 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MSGGDRFVQT LQKLNYPKGA QLDGEDFDWL FEAVDLKPFL DWFCSAASEQ NVVPDEKLQA FNTLKESGKP VLDEKALDEV LKTFSISKV PAIEEVAIEK LEEEVKALQK QKNLHIRRRN KLQMVESGNR QMCLKSKDKE EETGRAFQEV LHLLRVTNKK L NHELQSIV ...String: MSGGDRFVQT LQKLNYPKGA QLDGEDFDWL FEAVDLKPFL DWFCSAASEQ NVVPDEKLQA FNTLKESGKP VLDEKALDEV LKTFSISKV PAIEEVAIEK LEEEVKALQK QKNLHIRRRN KLQMVESGNR QMCLKSKDKE EETGRAFQEV LHLLRVTNKK L NHELQSIV NGVQTLMSFF STPETACELS SQPIFLSQLL LDKYLSLEEQ STAALTSFTK EHFFEGMSKF VEGSDENFQL VQ LNVNSFG EDGTTEDKCK EMMRLQLAYI CAKHKLIQMK AKSASLKVGL QWAENNASVV QDKASQKEEN LKVRITSLKN ETL QIENHT NSISNEKLPG LVRDNAQLLN MPIVKGDYDL QMAHQTSCSS RQDLVCDHLM KQKASFELLQ LGYELELRKH RDVY RELGS IVQELKESGD KLEERLTMLS DVNLLSASKP RSNIDSKDLT SHRLYQLLDG DNTQKLFRTY DGLESVAQKL SQDIA SMRD QLEVSEQEHS LLLSKLDSHL KELRDFMYPE GNTLMLTTPE LSGEFHQLGS QLEKLNHITV EILGDLQLKR KMLESN KLQ QIEKQLYVYF FQNEEQLKSI VGKLEAQTGG GSSA UniProtKB: HAUS augmin-like complex subunit 3 |
-Macromolecule #3: HAUS augmin like complex subunit 4 L homeolog
| Macromolecule | Name: HAUS augmin like complex subunit 4 L homeolog / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 41.256438 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MAQTLQYVSS RLSMLQIDEE DLERNAQFGK VLIELCPLLG PNGGSANLNR ELEETRRELL LQRKMWMRSE VIYQLVQEML LDLQVRKLE GSLTEEERKF QDGLQQCMLV SECSRLLTAD SVPPSDSTSI LGLDKQDLLD LLPPNMLVLW VRDRLQKQLE E ALKKKCFT ...String: MAQTLQYVSS RLSMLQIDEE DLERNAQFGK VLIELCPLLG PNGGSANLNR ELEETRRELL LQRKMWMRSE VIYQLVQEML LDLQVRKLE GSLTEEERKF QDGLQQCMLV SECSRLLTAD SVPPSDSTSI LGLDKQDLLD LLPPNMLVLW VRDRLQKQLE E ALKKKCFT FLSFHQPETD EEGDVLRAAK VLRLASTLED EKRRLQNEQE KHQEMRALLE KQQEIYPHVL LRCLSLLRQA AS ELRLRAQ SDIDRINAEY LEAKSNALFL KLRMEELQVL TDCYTPEKVL VHRQIRDTLE AGVKKEKQEL STSRQILSSY EFL GPEFEG LVQEYTRLKD KIKDNRWMLQ ELSKSLP UniProtKB: HAUS augmin like complex subunit 4 L homeolog |
-Macromolecule #4: HAUS augmin-like complex subunit 5
| Macromolecule | Name: HAUS augmin-like complex subunit 5 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 77.357281 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MERRSLAQEL KKWAVEEMGL PAQKAPSEEM LQRLFIGQCG DIWKFIIRHI HSHRTVRKIE GNLLWYQQLQ HTEAQRTAEE EQQQRRKQL CKEILELRAE LHHLQEQIQT AEREIVGQDL NCERAQDLCR RSLLLRAFNK KREEECEALC QSNKKIQYRC E QLQEIRRA ...String: MERRSLAQEL KKWAVEEMGL PAQKAPSEEM LQRLFIGQCG DIWKFIIRHI HSHRTVRKIE GNLLWYQQLQ HTEAQRTAEE EQQQRRKQL CKEILELRAE LHHLQEQIQT AEREIVGQDL NCERAQDLCR RSLLLRAFNK KREEECEALC QSNKKIQYRC E QLQEIRRA SQREVMFSAV DPDLSSSTFL EPEVLRDVRE VCKLRFKFLR SLHDDSISSS VHPGKEDLRS LSHQQWMSMA EK VWNTHTP NHILAALERL TLNSTQELKK LQFSQAADLS KGPSCQLKEF SEPITQSRSC NESTHLDPQE TLPSFHSLIQ EGW ANSVKV SSELRRVQSQ AQALSEHLAE RIQEIHKKLS DGSEVSVLTR AAFDAELRCV ILRGCRDALM QECRMLQEEA AGKK QEMKL LQQQQQNIQE ACLLLDKKQK HIQILIKGNS SSKSQIRRSS VEAQKYVQDK LLPWPQEIIQ ESQRLQDSIQ KEVKH FSAI CLPALLKVST DGFNLLPSRE LSINRMSNTH APYYGIFKGI YESVRLPLYK APESVLSHVA DMKKQLFFLR SQLSSR SEA ISKTQRALQK NTNPDTDALL KSLSDHYSLE LDEMVPKMQR LIQQCEKHQE YGKEVQATVM DWWEQPVQLC LPSEERG GL TLRQWRERWT VAVTALQRAT GSRS UniProtKB: HAUS augmin-like complex subunit 5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.2 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 12615 / Average electron dose: 69.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8at2: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)