+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse Elongator complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   | |||||||||
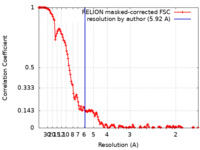
| Method | single particle reconstruction / cryo EM / Resolution: 5.92 Å | |||||||||
 Authors Authors | Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D ...Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D / Lin T-Y / Abbassi N / Hammermeister A / Chramiec-Glabik A / Kosinski J / Schaffrath R / Glatt S | |||||||||
| Funding support |  Poland, European Union, 2 items Poland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Cryo-EM structure of the fully assembled Elongator complex. Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej ...Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej Chramiec-Głąbik / Anna Biela / Dominika Dobosz / Ting-Yu Lin / Nour-El-Hana Abbassi / Alexander Hammermeister / Juri Rappsilber / Jan Kosinski / Raffael Schaffrath / Sebastian Glatt /    Abstract: Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition ...Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition of chemical groups. Modifications in the tRNA anticodons are directly influencing ribosome decoding and dynamics during translation elongation and are crucial for maintaining proteome integrity. In eukaryotes, wobble uridines are modified by Elongator, a large and highly conserved macromolecular complex. Elongator consists of two subcomplexes, namely Elp123 containing the enzymatically active Elp3 subunit and the associated Elp456 hetero-hexamer. The structure of the fully assembled complex and the function of the Elp456 subcomplex have remained elusive. Here, we show the cryo-electron microscopy structure of yeast Elongator at an overall resolution of 4.3 Å. We validate the obtained structure by complementary mutational analyses in vitro and in vivo. In addition, we determined various structures of the murine Elongator complex, including the fully assembled mouse Elongator complex at 5.9 Å resolution. Our results confirm the structural conservation of Elongator and its intermediates among eukaryotes. Furthermore, we complement our analyses with the biochemical characterization of the assembled human Elongator. Our results provide the molecular basis for the assembly of Elongator and its tRNA modification activity in eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15626.map.gz emd_15626.map.gz | 20 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15626-v30.xml emd-15626-v30.xml emd-15626.xml emd-15626.xml | 26.6 KB 26.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15626_fsc.xml emd_15626_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_15626.png emd_15626.png | 99.6 KB | ||
| Others |  emd_15626_half_map_1.map.gz emd_15626_half_map_1.map.gz emd_15626_half_map_2.map.gz emd_15626_half_map_2.map.gz | 225 MB 225.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15626 http://ftp.pdbj.org/pub/emdb/structures/EMD-15626 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15626 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15626 | HTTPS FTP |
-Validation report
| Summary document |  emd_15626_validation.pdf.gz emd_15626_validation.pdf.gz | 686.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15626_full_validation.pdf.gz emd_15626_full_validation.pdf.gz | 686.4 KB | Display | |
| Data in XML |  emd_15626_validation.xml.gz emd_15626_validation.xml.gz | 22.5 KB | Display | |
| Data in CIF |  emd_15626_validation.cif.gz emd_15626_validation.cif.gz | 29.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15626 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15626 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15626 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15626 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15626.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15626.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
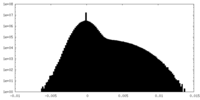
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
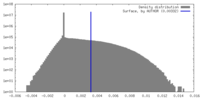
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15626_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
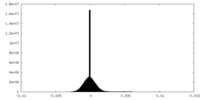
| Density Histograms |
-Half map: #1
| File | emd_15626_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
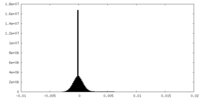
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse Elongator complex
| Entire | Name: Mouse Elongator complex |
|---|---|
| Components |
|
-Supramolecule #1: Mouse Elongator complex
| Supramolecule | Name: Mouse Elongator complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 828.2 KDa |
-Macromolecule #1: Elongator complex protein 1
| Macromolecule | Name: Elongator complex protein 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MRNLKLHRTL EFRDIQAPGK PQCFCLRAEQ GTVLIGSERG LTEVDPVRRE VKTEISLVAE GFLPEDGSG CIVGIQDLLD QESVCVATAS GDVIVCNLST QQLECVGSVA SGISVMSWSP D QELLLLAT AQQTLIMMTK DFEVIAEEQI HQDDFGEGKF VTVGWGSKQT ...String: MRNLKLHRTL EFRDIQAPGK PQCFCLRAEQ GTVLIGSERG LTEVDPVRRE VKTEISLVAE GFLPEDGSG CIVGIQDLLD QESVCVATAS GDVIVCNLST QQLECVGSVA SGISVMSWSP D QELLLLAT AQQTLIMMTK DFEVIAEEQI HQDDFGEGKF VTVGWGSKQT QFHGSEGRPT AF PVQLPEN ALPWDDRRPH ITWRGDGQYF AVSVVCRQTE ARKIRVWNRE FALQSTSESV PGL GPALAW KPSGSLIAST QDKPNQQDVV FFEKNGLLHG HFTLPFLKDE VKVNDLLWNA DSSV LAIWL EDLPKEDSST LKSYVQLWTV GNYHWYLKQS LPFSTTGKNQ IVSLLWDPVT PCRLH VLCT GWRYLCCDWH WTTDRSSGNS ANDLANVAVI DGNRVLVTVF RQTVVPPPMC TYRLLI PHP VNQVIFSAHL GNDLAVLDAS NQISVYKCGD KPNMDSTVKL GAVGGNGFKV PLTTPHL EK RYSIQFGNNE EEEEEEVNAL QLSFLTWVED DTFLAISYSH SSSQSIIHHL TVTHSEVD E EQGQLDVSSS VTVDGVVIGL CCCSKTKSLA VQLADGQVLK YLWESPSLAV EPWKNSEGI PVRFVHPCTQ MEVATIGGEE CVLGLTDRCR FFINDTEVAS NITSFAVCDD FLLVTTHSHT CQVFSLSGA SLKMLQAALS GSHEASGEIL RKVERGSRIV TVVPQDTKLI LQMPRGNLEV V HHRALVLA QIRKWLDKLM FKEAFECMRK LRINLNLIHD HNPKVFLENV ETFVKQIDSV NH INLFFTE LREEDVTKTM YPPPITKSVQ VSTHPDGKKL DLICDAMRAA MEAINPRKFC LSI LTSHVK KTTPELEIVL QKVQELQGNL PFDPESVSVE EALKYLLLLV DVNELFNHSL GTYD FNLVL MVAEKSQKDP KEYLPFLNTL KKMETNYQRF TIDKYLKRYE KALGHLSKCG PEYFT ECLN LIKDKNLYKE ALKLYRPDSP QYQAVSMAYG EHLMQEHLYE PAGLVFARCG AQEKAL EAF LACGSWQQAL CVAAQLQMSK DKVAGLARTL AGKLVEQRKH SEAATVLEQY AQDYEEA VL LLLEGSAWEE ALRLVYKYDR VDIIETSVKP SILEAQKNYM DFLDSETATF IRHKNRLQ V VRALRRQAPQ VHVDHEVAHG PESDLFSETS SIMSGSEMSG RYSHSNSRIS ARSSKNRRK AERKKHSLKE GSPLEGLALL EALSEVVQSV EKLKDEVRAI LKVLFLFEFE EQAKELQRAF ESTLQLMER AVPEIWTPAG QQSSTTPVLG PSSTANSITA SYQQQKTCVP ALDAGVYMPP K MDPRSQWK LSLLE |
-Macromolecule #2: Elongator complex protein 2
| Macromolecule | Name: Elongator complex protein 2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MVSSVLEVSH VFCCPNRVRG ALSWNTGPGG LLAFGTSCSV VLYDPQKKVV ITNLNGHTAR VNCLQWIRT EDGSPSNELV SGGSDNRVIH WELENNQVLK SVRLQGHEGP VCAVHAIYQS G PSEGEQHA LIASAASDST VRIWSKKGSE VKYLQTLSFR DGFVLSVCLA ...String: MVSSVLEVSH VFCCPNRVRG ALSWNTGPGG LLAFGTSCSV VLYDPQKKVV ITNLNGHTAR VNCLQWIRT EDGSPSNELV SGGSDNRVIH WELENNQVLK SVRLQGHEGP VCAVHAIYQS G PSEGEQHA LIASAASDST VRIWSKKGSE VKYLQTLSFR DGFVLSVCLA ILPGTNVPVL AC GDDDCRI HLYIQQDDQF QKALSLCGHE DWIRGVEWAT FGRDLFLASC SQDCLIRIWR LYM KPASFE TKDGSLRLKE NTFTIKDGGV RTTVAVTLET VLAGHENWVN AVHWQPSFYK DGVL QQPVR LLSASMDKTM ILWAPDEESG VWLEQVRVGE VGGNTLGFYD CQFGENGTMI IAHAF HGAL HLWKQSTVNP RQWAPEIVIS GHFDGVQDLM WDPEGEFIIT TSTDQTTRLF APWKKK DQK DRSQVTWHEI ARPQIHGYNI KCLAMIDRFQ FVSGADEKVL RVFSAPRNFV ENFSVIS RQ SLSHMLCDDQ DLPEGATVPA LGLSNKALFQ GDIASQPFEE DELISPAFGS PQVTFQPA V LNEPPTEDHL LQNTLWPEIQ KLYGHGYEIV CVACNNSKTL LASACKASQK EHAAIILWS TASWKQVQSL AFHTLTVTQM TFSPDDKFLL AVSRDRTWSL WKRQDATSSE FDPFFSLFAF TNKITSVHS RIIWSCDWSP DNKYFFTGSR DKKVVVWGEC KSSHNPMEHP IRPCSSILDV G SSVTAVSV CPVLNPAQRY IVAIGLESGK ICIYSWNKTN QEINDWTSCV ETNPSQSHSL GI RRLCWKS CSDDDDDDDD DDTEQSEEGP EWLHFASCGE DHTVKIYRVN RRAL |
-Macromolecule #3: Elongator complex protein 3
| Macromolecule | Name: Elongator complex protein 3 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MRQKRKGDLS PAELMMLTIG DVIKQLVEAH EQGKDVDLNK MKTKTAAKYG LASQPRLVDI IAAVPPHYR KILIPKLKAK PVRTASGIAV VAVMCKPHRC PHISFTGNIC IYCPGGPDSD F EYSTQSYT GYEPTSMRAI RARYDPFLQT RHRIEQLKQL GHSVDKVEFI ...String: MRQKRKGDLS PAELMMLTIG DVIKQLVEAH EQGKDVDLNK MKTKTAAKYG LASQPRLVDI IAAVPPHYR KILIPKLKAK PVRTASGIAV VAVMCKPHRC PHISFTGNIC IYCPGGPDSD F EYSTQSYT GYEPTSMRAI RARYDPFLQT RHRIEQLKQL GHSVDKVEFI VMGGTFMALP EE YRDYFIR SLHDALSGHT SNNIHEAIKY SERSFTKCVG ITIETRPDYC MKRHLSDMLT YGC TRLEIG VQSVYEDVAR DTNRGHTVKA ACESFHLAKD SGFKVVTHMM PDLPNVGLER DIEQ FIEFF ENPAFRPDGL KLYPTLVIRG TGLYELWKSG RYRSYSPSDL IELVARILAL VPPWT RVYR VQRDIPMPLV SSGVEHGNLR ELAFARMKDL GIQCRDVRTR EVGIQEIHHR VRPYQV ELV RRDYVANGGW ETFLSYEDPD QDILIGLLRL RKCSEETFRF ELGGGVSIVR ELHVYGS VV PVSSRDPTKF QHQGFGMLLM EEAERIAREE HGSGKMAVIS GVGTRNYYRK IGYRLQGP Y MVKMLK |
-Macromolecule #4: Elongator complex protein 4
| Macromolecule | Name: Elongator complex protein 4 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAAADTCGAG TLSSRSVASE AGQGGTSSFQ RKGKASGGPG GGPRLLSIAG TRPSVRNGQL LVSTGLPAL DQLLGGGLAV GTLLLIEEDK YNIYSPLLFK YFMAEGIING HTLLVASAKE N PAKILQEL PAPLLDDNSK KELEDVHSAK TPEPNVNMKI AWRYQLQPKM ...String: MAAADTCGAG TLSSRSVASE AGQGGTSSFQ RKGKASGGPG GGPRLLSIAG TRPSVRNGQL LVSTGLPAL DQLLGGGLAV GTLLLIEEDK YNIYSPLLFK YFMAEGIING HTLLVASAKE N PAKILQEL PAPLLDDNSK KELEDVHSAK TPEPNVNMKI AWRYQLQPKM EVGPVSSSRF GH YYDLSKR IPWELLQSSK WHGFFLPEHI SPDLKGESCF LSCGYMRLLE FIQKSVYAEG FDG ANPQKK QKNILRIGIQ NLGSPLWGDD ICCKENCDNN HRLTKFLYIL RGLLRSSLSA CIIT MPAHL VQNKSITTRV RNLSDTVVGL ESFIGSERET NPLYKDYHGL IHIRKIPRLN NLTCD ESDV KDLAFKLKRK LFTIERLHLP PDLSDTVGRS SKQDLAASTA RLGAGCSSMA EGKKHL DF |
-Macromolecule #5: Elongator complex protein 5
| Macromolecule | Name: Elongator complex protein 5 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MLDSLLAIGG LVLLRDSVEW EGRSLLKALI KKSALRGEQV HVLGCEVSEE EFREGFDSDV NSRLVYHDL FRDPLNWSKP GEAVPEGPLK ALRSMCKRTD HGSVTIALDS LSWLLCHIPC V TLCQALHA LSQQNGDPGD NSLVEQVRVL GLLHEELHGP GSMGALNTLA ...String: MLDSLLAIGG LVLLRDSVEW EGRSLLKALI KKSALRGEQV HVLGCEVSEE EFREGFDSDV NSRLVYHDL FRDPLNWSKP GEAVPEGPLK ALRSMCKRTD HGSVTIALDS LSWLLCHIPC V TLCQALHA LSQQNGDPGD NSLVEQVRVL GLLHEELHGP GSMGALNTLA HTEVTLSGKV DQ TSASILC RRPQQRATYQ TWWFSVLPDF SLTLHEGLPL RSELHPDHHT TQVDPTAHLT FNL HLSKKE REARDSLTLP FQFSSEKQKA LLHPVPSRTT GHIFYEPDAF DDVDPEDPDD DLDI |
-Macromolecule #6: Elongator complex protein 6
| Macromolecule | Name: Elongator complex protein 6 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MFPELNNLLS TTPDKTEQGT LTLLCDAKTD GSFLVHHFLS FYLKANCKVC FVALVQSFSH YNIVGQKLG VSLTAARDRG QLVFLEGLKS SVEVLFHSQD EPHPLQFLRE AGTGNLQSLY T FIQDTLKP ADSEESPWKY PVLLVDNLSV LLSLGVGAVA VLDFMQYCRA ...String: MFPELNNLLS TTPDKTEQGT LTLLCDAKTD GSFLVHHFLS FYLKANCKVC FVALVQSFSH YNIVGQKLG VSLTAARDRG QLVFLEGLKS SVEVLFHSQD EPHPLQFLRE AGTGNLQSLY T FIQDTLKP ADSEESPWKY PVLLVDNLSV LLSLGVGAVA VLDFMQYCRA TVCCELKGNV VA LVHDTEG ATDEGNDTLL NGLSHQSHLI LRAEGLATGF CKDVHGQLSI LWRRPSRSTA QRA QSLTYQ YKIQDKNVSF FAKGMSPAVL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 15 s wait time, blot force 5, 5 s blot time. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 5232 / Average exposure time: 1.82 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)