+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse heavy chain apoferritin in plunge-frozen vitreous ice | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.47 Å | |||||||||
 Authors Authors | Bongiovanni G / Harder OF / Drabbels M / Lorenz UJ | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Front Mol Biosci / Year: 2022 Journal: Front Mol Biosci / Year: 2022Title: Microsecond melting and revitrification of cryo samples with a correlative light-electron microscopy approach. Authors: Gabriele Bongiovanni / Oliver F Harder / Marcel Drabbels / Ulrich J Lorenz /  Abstract: We have recently introduced a novel approach to time-resolved cryo-electron microscopy (cryo-EM) that affords microsecond time resolution. It involves melting a cryo sample with a laser beam to allow ...We have recently introduced a novel approach to time-resolved cryo-electron microscopy (cryo-EM) that affords microsecond time resolution. It involves melting a cryo sample with a laser beam to allow dynamics of the embedded particles to occur. Once the laser beam is switched off, the sample revitrifies within just a few microseconds, trapping the particles in their transient configurations, which can subsequently be imaged to obtain a snap shot of the dynamics at this point in time. While we have previously performed such experiments with a modified transmission electron microscope, we here demonstrate a simpler implementation that uses an optical microscope. We believe that this will make our technique more easily accessible and hope that it will encourage other groups to apply microsecond time-resolved cryo-EM to study the fast dynamics of a variety of proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15619.map.gz emd_15619.map.gz | 779 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15619-v30.xml emd-15619-v30.xml emd-15619.xml emd-15619.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15619.png emd_15619.png | 184.4 KB | ||
| Others |  emd_15619_half_map_1.map.gz emd_15619_half_map_1.map.gz emd_15619_half_map_2.map.gz emd_15619_half_map_2.map.gz | 763 MB 763 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15619 http://ftp.pdbj.org/pub/emdb/structures/EMD-15619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15619 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15619.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15619.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.455 Å | ||||||||||||||||||||||||||||||||||||
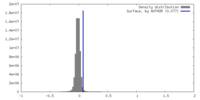
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15619_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
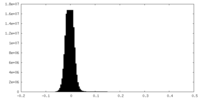
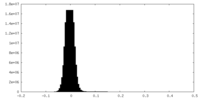
| Density Histograms |
-Half map: #2
| File | emd_15619_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
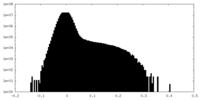
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse heavy-chain apoferritin
| Entire | Name: Mouse heavy-chain apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: Mouse heavy-chain apoferritin
| Supramolecule | Name: Mouse heavy-chain apoferritin / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 1.47 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 447704 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)