[English] 日本語
 Yorodumi
Yorodumi- EMDB-15566: rotational state 1d of the Trypanosoma brucei mitochondrial ATP s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | rotational state 1d of the Trypanosoma brucei mitochondrial ATP synthase dimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationH+-transporting two-sector ATPase / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances / kinetoplast / ATP biosynthetic process / nuclear lumen / ciliary plasm / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton motive force-driven mitochondrial ATP synthesis / proton-transporting ATPase activity, rotational mechanism ...H+-transporting two-sector ATPase / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances / kinetoplast / ATP biosynthetic process / nuclear lumen / ciliary plasm / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton motive force-driven mitochondrial ATP synthesis / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / proton transmembrane transport / ADP binding / mitochondrial membrane / mitochondrial inner membrane / hydrolase activity / lipid binding / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
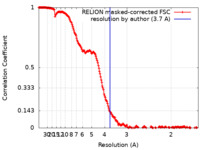
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Muehleip A / Gahura O / Zikova A / Amunts A | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases. Authors: Ondřej Gahura / Alexander Mühleip / Carolina Hierro-Yap / Brian Panicucci / Minal Jain / David Hollaus / Martina Slapničková / Alena Zíková / Alexey Amunts /   Abstract: Mitochondrial ATP synthase forms stable dimers arranged into oligomeric assemblies that generate the inner-membrane curvature essential for efficient energy conversion. Here, we report cryo-EM ...Mitochondrial ATP synthase forms stable dimers arranged into oligomeric assemblies that generate the inner-membrane curvature essential for efficient energy conversion. Here, we report cryo-EM structures of the intact ATP synthase dimer from Trypanosoma brucei in ten different rotational states. The model consists of 25 subunits, including nine lineage-specific, as well as 36 lipids. The rotary mechanism is influenced by the divergent peripheral stalk, conferring a greater conformational flexibility. Proton transfer in the lumenal half-channel occurs via a chain of five ordered water molecules. The dimerization interface is formed by subunit-g that is critical for interactions but not for the catalytic activity. Although overall dimer architecture varies among eukaryotes, we find that subunit-g together with subunit-e form an ancestral oligomerization motif, which is shared between the trypanosomal and mammalian lineages. Therefore, our data defines the subunit-g/e module as a structural component determining ATP synthase oligomeric assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15566.map.gz emd_15566.map.gz | 380.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15566-v30.xml emd-15566-v30.xml emd-15566.xml emd-15566.xml | 41.3 KB 41.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15566_fsc.xml emd_15566_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15566.png emd_15566.png | 84 KB | ||
| Masks |  emd_15566_msk_1.map emd_15566_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Others |  emd_15566_half_map_1.map.gz emd_15566_half_map_1.map.gz emd_15566_half_map_2.map.gz emd_15566_half_map_2.map.gz | 544 MB 544.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15566 http://ftp.pdbj.org/pub/emdb/structures/EMD-15566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15566 | HTTPS FTP |
-Related structure data
| Related structure data |  8apdMC  8ap6C  8ap7C  8ap8C  8ap9C  8apaC  8apbC  8apcC  8apeC  8apfC  8apgC  8aphC  8apjC  8apkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15566.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15566.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
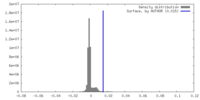
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15566_msk_1.map emd_15566_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
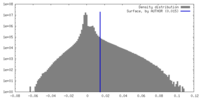
| Density Histograms |
-Half map: #1
| File | emd_15566_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
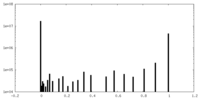
| Density Histograms |
-Half map: #2
| File | emd_15566_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
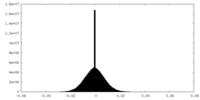
| Density Histograms |
- Sample components
Sample components
+Entire : mitochondrial ATP synthase dimer from Trypanosoma brucei
+Supramolecule #1: mitochondrial ATP synthase dimer from Trypanosoma brucei
+Macromolecule #1: subunit-e
+Macromolecule #2: subunit-g
+Macromolecule #3: ATP synthase subunit a
+Macromolecule #4: subunit-8
+Macromolecule #5: subunit-d
+Macromolecule #6: ATPTB1
+Macromolecule #7: subunit-f
+Macromolecule #8: ATPTB3
+Macromolecule #9: ATPTB4
+Macromolecule #10: subunit-i/j
+Macromolecule #11: ATPTB6
+Macromolecule #12: subunit-k
+Macromolecule #13: ATPTB11
+Macromolecule #14: ATPTB12
+Macromolecule #15: subunit-b
+Macromolecule #16: ATPEG3
+Macromolecule #17: ATPEG4
+Macromolecule #18: ATP synthase subunit alpha, mitochondrial
+Macromolecule #19: ATP synthase subunit beta, mitochondrial
+Macromolecule #20: ATP synthase gamma subunit
+Macromolecule #21: ATP synthase, epsilon chain, putative
+Macromolecule #22: ATP synthase subunit epsilon, mitochondrial
+Macromolecule #23: ATP synthase subunit p18, mitochondrial
+Macromolecule #24: OSCP
+Macromolecule #25: ATPase subunit 9, putative
+Macromolecule #26: CARDIOLIPIN
+Macromolecule #27: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
+Macromolecule #28: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #29: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #30: 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl...
+Macromolecule #31: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #32: MAGNESIUM ION
+Macromolecule #33: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #34: URIDINE 5'-TRIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)