+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
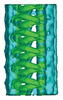
| Title | TAILS structure in human sperm tip singlet microtubules | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Microtubule Sperm TAILS / UNKNOWN FUNCTION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 40.0 Å | |||||||||
 Authors Authors | Hoog JL / Zabeo DZ | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2018 Journal: Sci Rep / Year: 2018Title: A lumenal interrupted helix in human sperm tail microtubules. Authors: Davide Zabeo / John M Heumann / Cindi L Schwartz / Azusa Suzuki-Shinjo / Garry Morgan / Per O Widlund / Johanna L Höög /   Abstract: Eukaryotic flagella are complex cellular extensions involved in many human diseases gathered under the term ciliopathies. Currently, detailed insights on flagellar structure come mostly from studies ...Eukaryotic flagella are complex cellular extensions involved in many human diseases gathered under the term ciliopathies. Currently, detailed insights on flagellar structure come mostly from studies on protists. Here, cryo-electron tomography (cryo-ET) was performed on intact human spermatozoon tails and showed a variable number of microtubules in the singlet region (inside the end-piece). Inside the microtubule plus end, a novel left-handed interrupted helix which extends several micrometers was discovered. This structure was named Tail Axoneme Intra-Lumenal Spiral (TAILS) and binds directly to 11 protofilaments on the internal microtubule wall, in a coaxial fashion with the surrounding microtubule lattice. It leaves a gap over the microtubule seam, which was directly visualized in both singlet and doublet microtubules. We speculate that TAILS may stabilize microtubules, enable rapid swimming or play a role in controlling the swimming direction of spermatozoa. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15450.map.gz emd_15450.map.gz | 695.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15450-v30.xml emd-15450-v30.xml emd-15450.xml emd-15450.xml | 7.9 KB 7.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15450.png emd_15450.png | 125 KB | ||
| Filedesc metadata |  emd-15450.cif.gz emd-15450.cif.gz | 3.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15450 http://ftp.pdbj.org/pub/emdb/structures/EMD-15450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15450 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15450.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15450.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.7 Å | ||||||||||||||||||||||||||||||||||||
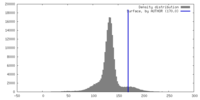
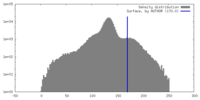
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Human sperm flagellum end piece
| Entire | Name: Human sperm flagellum end piece |
|---|---|
| Components |
|
-Supramolecule #1: Human sperm flagellum end piece
| Supramolecule | Name: Human sperm flagellum end piece / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Ejaculate applied directly to EM grid after collection. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 1.23 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 40.0 Å / Resolution method: OTHER / Number subtomograms used: 1940 |
|---|---|
| Extraction | Number tomograms: 2 / Number images used: 1940 |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)