+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of membrane-tethered poliovirus | ||||||||||||
 Map data Map data | Subtomogram average of membrane-tethered poliovirus | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | poliovirus / cellular tomography / in situ / VIRUS | ||||||||||||
| Biological species |   Human poliovirus 1 Mahoney Human poliovirus 1 Mahoney | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 70.0 Å | ||||||||||||
 Authors Authors | Dahmane S / Carlson LA | ||||||||||||
| Funding support |  France, European Union, France, European Union,  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Membrane-assisted assembly and selective secretory autophagy of enteroviruses. Authors: Selma Dahmane / Adeline Kerviel / Dustin R Morado / Kasturika Shankar / Björn Ahlman / Michael Lazarou / Nihal Altan-Bonnet / Lars-Anders Carlson /    Abstract: Enteroviruses are non-enveloped positive-sense RNA viruses that cause diverse diseases in humans. Their rapid multiplication depends on remodeling of cytoplasmic membranes for viral genome ...Enteroviruses are non-enveloped positive-sense RNA viruses that cause diverse diseases in humans. Their rapid multiplication depends on remodeling of cytoplasmic membranes for viral genome replication. It is unknown how virions assemble around these newly synthesized genomes and how they are then loaded into autophagic membranes for release through secretory autophagy. Here, we use cryo-electron tomography of infected cells to show that poliovirus assembles directly on replication membranes. Pharmacological untethering of capsids from membranes abrogates RNA encapsidation. Our data directly visualize a membrane-bound half-capsid as a prominent virion assembly intermediate. Assembly progression past this intermediate depends on the class III phosphatidylinositol 3-kinase VPS34, a key host-cell autophagy factor. On the other hand, the canonical autophagy initiator ULK1 is shown to restrict virion production since its inhibition leads to increased accumulation of virions in vast intracellular arrays, followed by an increased vesicular release at later time points. Finally, we identify multiple layers of selectivity in virus-induced autophagy, with a strong selection for RNA-loaded virions over empty capsids and the segregation of virions from other types of autophagosome contents. These findings provide an integrated structural framework for multiple stages of the poliovirus life cycle. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15392.map.gz emd_15392.map.gz | 10.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15392-v30.xml emd-15392-v30.xml emd-15392.xml emd-15392.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15392.png emd_15392.png | 17.9 KB | ||
| Masks |  emd_15392_msk_1.map emd_15392_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15392.cif.gz emd-15392.cif.gz | 4 KB | ||
| Others |  emd_15392_half_map_1.map.gz emd_15392_half_map_1.map.gz emd_15392_half_map_2.map.gz emd_15392_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15392 http://ftp.pdbj.org/pub/emdb/structures/EMD-15392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15392 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15392.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15392.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of membrane-tethered poliovirus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.29 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15392_msk_1.map emd_15392_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
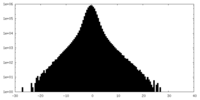
| Density Histograms |
-Half map: half map 1
| File | emd_15392_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
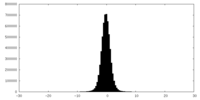
| Density Histograms |
-Half map: half map 2
| File | emd_15392_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human poliovirus 1 Mahoney
| Entire | Name:   Human poliovirus 1 Mahoney Human poliovirus 1 Mahoney |
|---|---|
| Components |
|
-Supramolecule #1: Human poliovirus 1 Mahoney
| Supramolecule | Name: Human poliovirus 1 Mahoney / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 12081 / Sci species name: Human poliovirus 1 Mahoney / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 3.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 70.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Dynamo / Number subtomograms used: 179 |
|---|---|
| Extraction | Number tomograms: 22 / Number images used: 241 |
| Final 3D classification | Number classes: 1 / Avg.num./class: 1 |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: Dynamo |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)