+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of RecF-RecR-DNA from Thermus thermophilus. | |||||||||
 Map data Map data | Sharpened Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA binding protein / DNA Repair pathway / RecFOR pathway / Thermus thermophilus | |||||||||
| Function / homology |  Function and homology information Function and homology informationSOS response / DNA synthesis involved in DNA repair / double-strand break repair / single-stranded DNA binding / DNA recombination / DNA replication / DNA repair / DNA binding / zinc ion binding / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) / synthetic construct (others) Thermus thermophilus HB8 (bacteria) / synthetic construct (others) | |||||||||
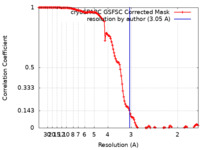
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Nirwal S / Czarnocki-Cieciura M / Chaudhary A / Zajko W / Skowronek K / Chamera S / Figiel M / Nowotny M | |||||||||
| Funding support |  Poland, 1 items Poland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination. Authors: Shivlee Nirwal / Mariusz Czarnocki-Cieciura / Anuradha Chaudhary / Weronika Zajko / Krzysztof Skowronek / Sebastian Chamera / Małgorzata Figiel / Marcin Nowotny /  Abstract: In bacteria, one type of homologous-recombination-based DNA-repair pathway involves RecFOR proteins that bind at the junction between single-stranded (ss) and double-stranded (ds) DNA. They ...In bacteria, one type of homologous-recombination-based DNA-repair pathway involves RecFOR proteins that bind at the junction between single-stranded (ss) and double-stranded (ds) DNA. They facilitate the replacement of SSB protein, which initially covers ssDNA, with RecA, which mediates the search for homologous sequences. However, the molecular mechanism of RecFOR cooperation remains largely unknown. We used Thermus thermophilus proteins to study this system. Here, we present a cryo-electron microscopy structure of the RecF-dsDNA complex, and another reconstruction that shows how RecF interacts with two different regions of the tetrameric RecR ring. Lower-resolution reconstructions of the RecR-RecO subcomplex and the RecFOR-DNA assembly explain how RecO is positioned to interact with ssDNA and SSB, which is proposed to lock the complex on a ssDNA-dsDNA junction. Our results integrate the biochemical data available for the RecFOR system and provide a framework for its complete understanding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15267.map.gz emd_15267.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15267-v30.xml emd-15267-v30.xml emd-15267.xml emd-15267.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15267_fsc.xml emd_15267_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15267.png emd_15267.png | 107.6 KB | ||
| Masks |  emd_15267_msk_1.map emd_15267_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15267.cif.gz emd-15267.cif.gz | 6.9 KB | ||
| Others |  emd_15267_additional_1.map.gz emd_15267_additional_1.map.gz emd_15267_half_map_1.map.gz emd_15267_half_map_1.map.gz emd_15267_half_map_2.map.gz emd_15267_half_map_2.map.gz | 122.4 MB 226.8 MB 226.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15267 http://ftp.pdbj.org/pub/emdb/structures/EMD-15267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15267 | HTTPS FTP |
-Validation report
| Summary document |  emd_15267_validation.pdf.gz emd_15267_validation.pdf.gz | 969.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15267_full_validation.pdf.gz emd_15267_full_validation.pdf.gz | 969.3 KB | Display | |
| Data in XML |  emd_15267_validation.xml.gz emd_15267_validation.xml.gz | 22.2 KB | Display | |
| Data in CIF |  emd_15267_validation.cif.gz emd_15267_validation.cif.gz | 28.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15267 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15267 | HTTPS FTP |
-Related structure data
| Related structure data |  8a93MC  8a8jC  8ab0C  8bprC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15267.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15267.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15267_msk_1.map emd_15267_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Raw map
| File | emd_15267_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15267_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15267_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of RecF-RecR-DNA from Thermus thermophilus.
| Entire | Name: Complex of RecF-RecR-DNA from Thermus thermophilus. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of RecF-RecR-DNA from Thermus thermophilus.
| Supramolecule | Name: Complex of RecF-RecR-DNA from Thermus thermophilus. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: DNA replication and repair protein RecF
| Macromolecule | Name: DNA replication and repair protein RecF / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: ATCC 27634 / DSM 579 / HB8 Thermus thermophilus HB8 (bacteria) / Strain: ATCC 27634 / DSM 579 / HB8 |
| Molecular weight | Theoretical: 37.933715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMRLLLFRQR NFRNLALEAY RPPPGLSALV GANAQGKTSL LLGIHLALGG EVPLGLADLV RFGEEEAWLH AEVETELGAY RLEHRLGPG GREVLLNGKR VSLRTLWELP GSVLVSPLDL EAVLGPKEER RAYLDRLIAR FSRRYAALLS AYEKALRQRN A LLKAGGEG ...String: SMRLLLFRQR NFRNLALEAY RPPPGLSALV GANAQGKTSL LLGIHLALGG EVPLGLADLV RFGEEEAWLH AEVETELGAY RLEHRLGPG GREVLLNGKR VSLRTLWELP GSVLVSPLDL EAVLGPKEER RAYLDRLIAR FSRRYAALLS AYEKALRQRN A LLKAGGEG LSAWDRELAR YGDEIVALRR RFLRRFAPIL REVHAALAAK EAGLRLEETA GEGVLRALEA SRAEERERGQ TL VGPHRDD LVFLLEGRPA HRFASRGEAK TLALALRLAE HRLLGEHHGE PPLLLVDEWG EELDEARRRA VLAYAQALPQ AIL AGLEAP PGVPVCSVVR GVVLCPGA UniProtKB: DNA replication and repair protein RecF |
-Macromolecule #2: Recombination protein RecR
| Macromolecule | Name: Recombination protein RecR / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: ATCC 27634 / DSM 579 / HB8 Thermus thermophilus HB8 (bacteria) / Strain: ATCC 27634 / DSM 579 / HB8 |
| Molecular weight | Theoretical: 21.323467 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMRYPESLLK LTRALSRLPG IGPKTAQRLA LHLAFHKEEA EALAEALEGI KRVRACRECG NLAEGELCPI CQDEDRDRSL LAVVESVAD LYALERSGEF RGLYHVLGGA LNPLEGIGPK ELNLEGLFRR LEGVEEVVLA TSMTVEGEAT ALYLAEELKK R GVRVTRPA ...String: SMRYPESLLK LTRALSRLPG IGPKTAQRLA LHLAFHKEEA EALAEALEGI KRVRACRECG NLAEGELCPI CQDEDRDRSL LAVVESVAD LYALERSGEF RGLYHVLGGA LNPLEGIGPK ELNLEGLFRR LEGVEEVVLA TSMTVEGEAT ALYLAEELKK R GVRVTRPA YGLPVGGSLE YADEVTLGRA LEGRRPV UniProtKB: Recombination protein RecR |
-Macromolecule #3: Oligo1
| Macromolecule | Name: Oligo1 / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.661912 KDa |
| Sequence | String: (DG)(DG)(DC)(DC)(DA)(DG)(DA)(DT)(DC)(DT) (DG)(DC)(DC)(DG)(DC)(DG)(DG)(DA)(DT)(DC) (DC)(DG)(DC)(DG)(DC) |
-Macromolecule #4: Oligo2
| Macromolecule | Name: Oligo2 / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.395925 KDa |
| Sequence | String: (DG)(DC)(DG)(DC)(DG)(DG)(DA)(DT)(DC)(DC) (DG)(DC)(DG)(DG)(DC)(DA)(DG)(DA)(DT)(DC) (DT)(DG)(DG)(DC)(DC)(DT)(DG)(DA)(DT) (DT)(DG)(DC)(DG)(DG)(DT)(DA)(DC)(DA)(DG) (DA) |
-Macromolecule #5: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Sample fixed with 0.05% glutaraldehyde and concentrated prior to vitrification; exact concentration cannot be estimated accurately. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 7217 / Average electron dose: 41.71 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8a93: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)