+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PPSA C terminal octahedral structure | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | enzyme / phosphorylation of pyruvate / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate, water dikinase / pyruvate, water dikinase activity / gluconeogenesis / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Pyrococcus furiosus DSM 3638 (archaea) Pyrococcus furiosus DSM 3638 (archaea) | |||||||||
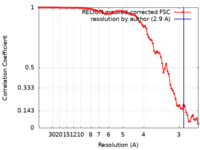
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Song W | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Survival strategies in the heat Lysine acetylation stabilizes the quaternary structure of a Mega-Dalton hyperthermophilic PEP-synthase Authors: Song W | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15230.map.gz emd_15230.map.gz | 7.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15230-v30.xml emd-15230-v30.xml emd-15230.xml emd-15230.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15230_fsc.xml emd_15230_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_15230.png emd_15230.png | 211.8 KB | ||
| Masks |  emd_15230_msk_1.map emd_15230_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15230.cif.gz emd-15230.cif.gz | 5.6 KB | ||
| Others |  emd_15230_half_map_1.map.gz emd_15230_half_map_1.map.gz emd_15230_half_map_2.map.gz emd_15230_half_map_2.map.gz | 36.8 MB 36.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15230 http://ftp.pdbj.org/pub/emdb/structures/EMD-15230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15230 | HTTPS FTP |
-Validation report
| Summary document |  emd_15230_validation.pdf.gz emd_15230_validation.pdf.gz | 687.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15230_full_validation.pdf.gz emd_15230_full_validation.pdf.gz | 686.9 KB | Display | |
| Data in XML |  emd_15230_validation.xml.gz emd_15230_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_15230_validation.cif.gz emd_15230_validation.cif.gz | 20.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15230 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15230 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15230 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15230 | HTTPS FTP |
-Related structure data
| Related structure data |  8a8eMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15230.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15230.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15230_msk_1.map emd_15230_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15230_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15230_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Phosphoenolpyruvate synthase
| Entire | Name: Phosphoenolpyruvate synthase |
|---|---|
| Components |
|
-Supramolecule #1: Phosphoenolpyruvate synthase
| Supramolecule | Name: Phosphoenolpyruvate synthase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus DSM 3638 (archaea) Pyrococcus furiosus DSM 3638 (archaea) |
| Molecular weight | Theoretical: 2.3 MDa |
-Macromolecule #1: Phosphoenolpyruvate synthase
| Macromolecule | Name: Phosphoenolpyruvate synthase / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO / EC number: pyruvate, water dikinase |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus DSM 3638 (archaea) / Strain: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus DSM 3638 (archaea) / Strain: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| Molecular weight | Theoretical: 90.600922 KDa |
| Recombinant expression | Organism:   Pyrococcus furiosus DSM 3638 (archaea) Pyrococcus furiosus DSM 3638 (archaea) |
| Sequence | String: MAYRFIKWFE ELSKNDVPLV GGKGANLGEM TNAGIPVPPG FCVTAEAYKY FVENVKVSKE DVKRILGEKV NKGTISEVLA QAPDEPRPL QDWIMDIISK TDVDDSKMLQ ENTEAIRTLI KSLDMPSEIA EEIKQAYKEL SQRFGQEEVY VAVRSSATAE D LPEASFAG ...String: MAYRFIKWFE ELSKNDVPLV GGKGANLGEM TNAGIPVPPG FCVTAEAYKY FVENVKVSKE DVKRILGEKV NKGTISEVLA QAPDEPRPL QDWIMDIISK TDVDDSKMLQ ENTEAIRTLI KSLDMPSEIA EEIKQAYKEL SQRFGQEEVY VAVRSSATAE D LPEASFAG QQETYLDVLG ADDVIDKVKR CWASLWTARA TFYRAKQGFD HSKVYLSAVV QKMVNSEKSG VMFTANPVTN NR NEIMINA SWGLGEAVVS GAVTPDEYIV EKGTWKIKEK VIAKKEVMVI RNPETGRGTV MVKVAEYLGP EWVEKQVLTD EQI IEVAKM GQKIEDHYGW PQDIEWAYDK DDGKLYIVQS RPITTLKEEA TAEEAEEVEE AEVILKGLGA SPGIGAGRVV VIFD ASEID KVKEGDILVT TMTNPDMVPA MKRAAAIVTD EGGRTSHAAI VSRELGIPCV VGTKEATKKL KTGMYVTVDG TRGLV YKGI VKSLVKKKEE AKAEGGQVVV AGAPLVTGTM VKVNVSMPEV AERAAATGAD GVGLLRAEHM ILSIGQHPIK FIKEGK EEE LVEKLAEGIE KVAAAFYPRP VWYRTLDAPT NEFREMPGGE DEPEERNPML GWRGIRRGLD QPELLRAEFK AIKKVVE KG YNNIGVMLPL VSHPEQIREA KRIAREVGLE PHKDVAWGVM IEVPAAAIII EDLIKEGIDF VSFGTNDLTQ YTLAIDRD N ERVAKLYDET HPAVLKLIKH VIKVCKRYGV ETSICGQAGS DPKMARILVR LGIDSISANP DAVQLIRQVV AQEERKLML EAARKQLFEE EEEEELF UniProtKB: Phosphoenolpyruvate synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Sugar embedding | Material: Ice Details: The sample was plunged freeze in ethane-propane and transfer to liquid nitrogen. | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 298.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-8a8e: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)