[English] 日本語
 Yorodumi
Yorodumi- EMDB-15229: T5 phage receptor-binding protein pb5 bound to ferrichrome transp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA | |||||||||
 Map data Map data | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA at 3.1 angstroms | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacteriophage / receptor / TonB-dependent transporter / outer membrane / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsiderophore transmembrane transport / siderophore-iron import into cell / siderophore uptake transmembrane transporter activity / virus tail / transmembrane transporter complex / virion binding / toxic substance binding / cell outer membrane / signaling receptor activity / intracellular iron ion homeostasis ...siderophore transmembrane transport / siderophore-iron import into cell / siderophore uptake transmembrane transporter activity / virus tail / transmembrane transporter complex / virion binding / toxic substance binding / cell outer membrane / signaling receptor activity / intracellular iron ion homeostasis / entry receptor-mediated virion attachment to host cell / receptor-mediated virion attachment to host cell / iron ion binding / protein domain specific binding / symbiont entry into host cell / membrane Similarity search - Function | |||||||||
| Biological species |  Escherichia phage T5 (virus) / Escherichia phage T5 (virus) /  | |||||||||
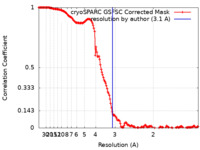
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Silale A / van den Berg B | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural basis for host recognition and superinfection exclusion by bacteriophage T5. Authors: Bert van den Berg / Augustinas Silale / Arnaud Baslé / Astrid F Brandner / Sophie L Mader / Syma Khalid /  Abstract: A key but poorly understood stage of the bacteriophage life cycle is the binding of phage receptor-binding proteins (RBPs) to receptors on the host cell surface, leading to injection of the phage ...A key but poorly understood stage of the bacteriophage life cycle is the binding of phage receptor-binding proteins (RBPs) to receptors on the host cell surface, leading to injection of the phage genome and, for lytic phages, host cell lysis. To prevent secondary infection by the same or a closely related phage and nonproductive phage adsorption to lysed cell fragments, superinfection exclusion (SE) proteins can prevent the binding of RBPs via modulation of the host receptor structure in ways that are also unclear. Here, we present the cryogenic electron microscopy (cryo-EM) structure of the phage T5 outer membrane (OM) receptor FhuA in complex with the T5 RBP pb5, and the crystal structure of FhuA complexed to the OM SE lipoprotein Llp. Pb5 inserts four loops deeply into the extracellular lumen of FhuA and contacts the plug but does not cause any conformational changes in the receptor, supporting the view that DNA translocation does not occur through the lumen of OM channels. The FhuA-Llp structure reveals that Llp is periplasmic and binds to a nonnative conformation of the plug of FhuA, causing the inward folding of two extracellular loops via "reverse" allostery. The inward-folded loops of FhuA overlap with the pb5 binding site, explaining how Llp binding to FhuA abolishes further infection of by phage T5 and suggesting a mechanism for SE via the jamming of TonB-dependent transporters by small phage lipoproteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15229.map.gz emd_15229.map.gz | 306.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15229-v30.xml emd-15229-v30.xml emd-15229.xml emd-15229.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15229_fsc.xml emd_15229_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_15229.png emd_15229.png | 45.9 KB | ||
| Masks |  emd_15229_msk_1.map emd_15229_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15229.cif.gz emd-15229.cif.gz | 6.9 KB | ||
| Others |  emd_15229_half_map_1.map.gz emd_15229_half_map_1.map.gz emd_15229_half_map_2.map.gz emd_15229_half_map_2.map.gz | 301.5 MB 301.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15229 http://ftp.pdbj.org/pub/emdb/structures/EMD-15229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15229 | HTTPS FTP |
-Related structure data
| Related structure data |  8a8cMC  8a60C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15229.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15229.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA at 3.1 angstroms | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15229_msk_1.map emd_15229_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: T5 phage receptor-binding protein pb5 bound to ferrichrome...
| File | emd_15229_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA at 3.1 angstroms, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: T5 phage receptor-binding protein pb5 bound to ferrichrome...
| File | emd_15229_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA at 3.1 angstroms, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : T5 phage receptor-binding protein pb5 bound to ferrichrome transp...
| Entire | Name: T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA |
|---|---|
| Components |
|
-Supramolecule #1: T5 phage receptor-binding protein pb5 bound to ferrichrome transp...
| Supramolecule | Name: T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 148 KDa |
-Macromolecule #1: Ferrichrome outer membrane transporter/phage receptor
| Macromolecule | Name: Ferrichrome outer membrane transporter/phage receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.816859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AVEPKEDTIT VTAAPAPQES AWGPAATIAA RQSATGTKTD TPIQKVPQSI SVVTAEEMAL HQPKSVKEAL SYTPGVSVGT RGASNTYDH LIIRGFAAEG QSQNNYLNGL KLQGNFYNDA VIDPYMLERA EIMRGPVSVL YGKSSPGGLL NMVSKRPTTE P LKEVQFKA ...String: AVEPKEDTIT VTAAPAPQES AWGPAATIAA RQSATGTKTD TPIQKVPQSI SVVTAEEMAL HQPKSVKEAL SYTPGVSVGT RGASNTYDH LIIRGFAAEG QSQNNYLNGL KLQGNFYNDA VIDPYMLERA EIMRGPVSVL YGKSSPGGLL NMVSKRPTTE P LKEVQFKA GTDSLFQTGF DFSDSLDDDG VYSYRLTGLA RSANAQQKGS EEQRYAIAPA FTWRPDDKTN FTFLSYFQNE PE TGYYGWL PKEGTVEPLP NGKRLPTDFN EGAKNNTYSR NEKMVGYSFD HEFNDTFTVR QNLRFAENKT SQNSVYGYGV CSD PANAYS KQCAALAPAD KGHYLARKYV VDDEKLQNFS VDTQLQSKFA TGDIDHTLLT GVDFMRMRND INAWFGYDDS VPLL NLYNP VNTDFDFNAK DPANSGPYRI LNKQKQTGVY VQDQAQWDKV LVTLGGRYDW ADQESLNRVA GTTDKRDDKQ FTWRG GVNY LFDNGVTPYF SYSESFEPSS QVGKDGNIFA PSKGKQYEVG VKYVPEDRPI VVTGAVYNLT KTNNLMADPE GSFFSV EGG EIRARGVEIE AKAALSASVN VVGSYTYTDA EYTTDTTYKG NTPAQVPKHM ASLWADYTFF DGPLSGLTLG TGGRYTG SS YGDPANSFKV GSYTVVDALV RYDLARVGMA GSNVALHVNN LFDREYVASC FNTYGCFWGA ERQVVATATF RF UniProtKB: Ferrichrome outer membrane transporter/phage receptor |
-Macromolecule #2: Receptor-binding protein pb5
| Macromolecule | Name: Receptor-binding protein pb5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 69.611453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFFAGKLNN KSILSLRRGS GGDTNQHINP DSQTIFHSDM SHVIITETHS TGLRLDQGAG DYYWSEMPSR VTQLHNNDPN RVVLTEIEF SDGSRHMLSG MSMGVGAKAY GIINPQIMSQ GGLKTQITAS ADLSLDVGYF NTGTSGTIPQ KLRDGTGCQH M FGAFSGRR ...String: MSFFAGKLNN KSILSLRRGS GGDTNQHINP DSQTIFHSDM SHVIITETHS TGLRLDQGAG DYYWSEMPSR VTQLHNNDPN RVVLTEIEF SDGSRHMLSG MSMGVGAKAY GIINPQIMSQ GGLKTQITAS ADLSLDVGYF NTGTSGTIPQ KLRDGTGCQH M FGAFSGRR GFASSAMYLG GAALYKSAWS GSGYVVADAG TLTIPSDYVR HPGARNFGFN AIYVRGRSCN RVLYGMEGPN YT TGGAVQG ASSSGALNFT YNPSNPESPK YSVGFARADP TNYAYWESMG DPNDSANGPI GIYSEHLGIY PSKITWYVTN LVY NGSGYN IDGGLFNGND IKLSPREFII KGVNVNNTSW KFINFIEKNF NVGNRADFRD VGCNLSKDSP STGISGIATF GLPT TESNN APSIKGGNVG GLHANVVSIY NFLPSASWYV SSNPPKIGNN YGDVWSENLL PLRLLGGSGS TILSGNIVFQ GNGSV HVGT VGLDLNSSRN GAIVCTMEFI DDTWLSAGGI GCFNPTEMLS QGAEYGDSRF RIGGNTINKK LHQILSLPAG EYVPFF TIK GTVVNACKLQ AAAYNPTPYW VSGLPGSVGQ TGYYTLTYYM RNDGNNNISI WLDSSMSNII GMKACLPNIK LIIQRLT HH HHHH UniProtKB: Receptor-binding protein pb5 |
-Macromolecule #3: [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethy...
| Macromolecule | Name: [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl] ...Name: [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate type: ligand / ID: 3 / Number of copies: 1 / Formula: LU9 |
|---|---|
| Molecular weight | Theoretical: 1.996235 KDa |
| Chemical component information |  ChemComp-LU9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 8387 / Average exposure time: 2.74 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)