[English] 日本語
 Yorodumi
Yorodumi- EMDB-15209: Nudaurelia capensis omega virus maturation intermediate captured ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | ||||||||||||
 Map data Map data | Postprocessed map from Relion | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ICOSAHEDRAL VIRUS / AUTO-CATALYTIC CLEAVAGE / VIRUS MATURATION / VIRUS-LIKE PARTICLE / VIRUS LIKE PARTICLE | ||||||||||||
| Function / homology | Peptidase N2 / Peptidase family A21 / virion component / Viral coat protein subunit / p70 Function and homology information Function and homology information | ||||||||||||
| Biological species |   Nudaurelia capensis omega virus Nudaurelia capensis omega virus | ||||||||||||
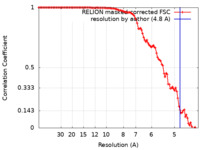
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | ||||||||||||
 Authors Authors | Castells-Graells R / Hesketh EL / Johnson JE / Ranson NA / Lawson DM / Lomonossoff GP | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Decoding virus maturation with cryo-EM structures of intermediates Authors: Castells-Graells R / Hesketh EL / Johnson JE / Ranson NA / Lawson DM / Lomonossoff GP | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15209.map.gz emd_15209.map.gz | 93.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15209-v30.xml emd-15209-v30.xml emd-15209.xml emd-15209.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15209_fsc.xml emd_15209_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_15209.png emd_15209.png | 193.4 KB | ||
| Masks |  emd_15209_msk_1.map emd_15209_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15209.cif.gz emd-15209.cif.gz | 6.2 KB | ||
| Others |  emd_15209_half_map_1.map.gz emd_15209_half_map_1.map.gz emd_15209_half_map_2.map.gz emd_15209_half_map_2.map.gz | 79.8 MB 80 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15209 http://ftp.pdbj.org/pub/emdb/structures/EMD-15209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15209 | HTTPS FTP |
-Validation report
| Summary document |  emd_15209_validation.pdf.gz emd_15209_validation.pdf.gz | 977.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15209_full_validation.pdf.gz emd_15209_full_validation.pdf.gz | 977.3 KB | Display | |
| Data in XML |  emd_15209_validation.xml.gz emd_15209_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_15209_validation.cif.gz emd_15209_validation.cif.gz | 23.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15209 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15209 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15209 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15209 | HTTPS FTP |
-Related structure data
| Related structure data |  8a6jMC  8aayC  8ac6C  8achC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15209.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15209.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map from Relion | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||
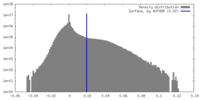
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15209_msk_1.map emd_15209_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refine3D half map 1 from Relion
| File | emd_15209_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D half map 1 from Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refine3D half map 2 from Relion
| File | emd_15209_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D half map 2 from Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nudaurelia capensis omega virus
| Entire | Name:   Nudaurelia capensis omega virus Nudaurelia capensis omega virus |
|---|---|
| Components |
|
-Supramolecule #1: Nudaurelia capensis omega virus
| Supramolecule | Name: Nudaurelia capensis omega virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: expressed in Spodoptera frugiperda Sf21 cells / NCBI-ID: 12541 / Sci species name: Nudaurelia capensis omega virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Gonimbrasia cytherea (butterflies/moths) Gonimbrasia cytherea (butterflies/moths) |
| Molecular weight | Theoretical: 16.76 MDa |
| Virus shell | Shell ID: 1 / Name: coat / Diameter: 460.0 Å / T number (triangulation number): 4 |
-Macromolecule #1: p70
| Macromolecule | Name: p70 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Nudaurelia capensis omega virus Nudaurelia capensis omega virus |
| Molecular weight | Theoretical: 69.891953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSNSASGKR RSRNVRIAAN TVNVAPKQRQ ARGRRARSRA NNIDNVTAAA QELGQSLDAN VITFPTNVAT MPEFRSWARG KLDIDQDSI GWYFKYLDPA GATESARAVG EYSKIPDGLV KFSVDAEIRE IYNEECPTVS DASIPLDGAQ WSLSIISYPM F RTAYFAVA ...String: MDSNSASGKR RSRNVRIAAN TVNVAPKQRQ ARGRRARSRA NNIDNVTAAA QELGQSLDAN VITFPTNVAT MPEFRSWARG KLDIDQDSI GWYFKYLDPA GATESARAVG EYSKIPDGLV KFSVDAEIRE IYNEECPTVS DASIPLDGAQ WSLSIISYPM F RTAYFAVA NVDNKEISLD VTNDLIVWLN NLASWRDVVD SGQWFTFSDD PTWFVRIRVL HPTYDLPDPT EGLLRTVSDY RL TYKSITC EANMPTLVDQ GFWIGGHYAL TPIATTQNAV EGSGFVHPFN VTRPGIAAGV TLTWASMPPG GSAPSGDPAW IPD STTQFQ WRHGGFDAPT GVITYTIPRG YTMQYFDTTT NEWNGFANPD DVVTFGQTGG AAGTNATITI TAPTVTLTIL ATTT SAANV INFRNLDAET TAASNRSEVP LPPLTFGQTA PNNPKIEQTL VKDTLGSYLV HSKMRNPVFQ LTPASSFGAI SFTNP GFDR NLDLPGFGGI RDSLDVNMST AVCHFRSLSK SCSIVTKTYQ GWEGVTNVNT PFGQFAHSGL LKNDEILCLA DDLATR LTG VYGATDNFAA AVLAFAANML TSVLKSEATT SVIKELGNQA TGLANQGLAR LPGLLASIPG KIAARVRARR DRRRAAR MN NN UniProtKB: p70 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 6.25 |
| Grid | Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 10528 / Average exposure time: 1.0 sec. / Average electron dose: 50.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 432 / Target criteria: Correlation coefficient |
| Output model |  PDB-8a6j: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)