[English] 日本語
 Yorodumi
Yorodumi- EMDB-15073: Rabies virus glycoprotein in complex with Fab fragments of 17C7 a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies | ||||||||||||||||||||||||
 Map data Map data | Sharpened final cryo-EM map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Viral Glycoprotein / Antibody / Complex / VIRAL PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral membrane / entry receptor-mediated virion attachment to host cell / fusion of virus membrane with host endosome membrane / viral envelope / virion membrane / membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Rabies lyssavirus / Rabies lyssavirus /  Rabies virus strain Pasteur vaccin / Rabies virus strain Pasteur vaccin /  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
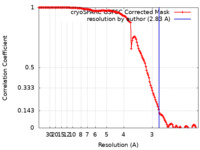
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | ||||||||||||||||||||||||
 Authors Authors | Ng WM / Fedosyuk S / English S / Augusto G / Berg A / Thorley L / Haselon AS / Segireddy RR / Bowden TA / Douglas AD | ||||||||||||||||||||||||
| Funding support |  United Kingdom, European Union, 7 items United Kingdom, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2022 Journal: Cell Host Microbe / Year: 2022Title: Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies. Authors: Weng M Ng / Sofiya Fedosyuk / Solomon English / Gilles Augusto / Adam Berg / Luke Thorley / Anna-Sophie Haselon / Rameswara R Segireddy / Thomas A Bowden / Alexander D Douglas /  Abstract: Rabies virus (RABV) causes lethal encephalitis and is responsible for approximately 60,000 deaths per year. As the sole virion-surface protein, the rabies virus glycoprotein (RABV-G) mediates host- ...Rabies virus (RABV) causes lethal encephalitis and is responsible for approximately 60,000 deaths per year. As the sole virion-surface protein, the rabies virus glycoprotein (RABV-G) mediates host-cell entry. RABV-G's pre-fusion trimeric conformation displays epitopes bound by protective neutralizing antibodies that can be induced by vaccination or passively administered for post-exposure prophylaxis. We report a 2.8-Å structure of a RABV-G trimer in the pre-fusion conformation, in complex with two neutralizing and protective monoclonal antibodies, 17C7 and 1112-1, that recognize distinct epitopes. One of these antibodies is a licensed prophylactic (17C7, Rabishield), which we show locks the protein in pre-fusion conformation. Targeted mutations can similarly stabilize RABV-G in the pre-fusion conformation, a key step toward structure-guided vaccine design. These data reveal the higher-order architecture of a key therapeutic target and the structural basis of neutralization by antibodies binding two key antigenic sites, and this will facilitate the development of improved vaccines and prophylactic antibodies. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15073.map.gz emd_15073.map.gz | 230.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15073-v30.xml emd-15073-v30.xml emd-15073.xml emd-15073.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15073_fsc.xml emd_15073_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15073.png emd_15073.png | 123.7 KB | ||
| Masks |  emd_15073_msk_1.map emd_15073_msk_1.map emd_15073_msk_2.map emd_15073_msk_2.map | 244.1 MB 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15073.cif.gz emd-15073.cif.gz | 7.2 KB | ||
| Others |  emd_15073_half_map_1.map.gz emd_15073_half_map_1.map.gz emd_15073_half_map_2.map.gz emd_15073_half_map_2.map.gz | 226.9 MB 226.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15073 http://ftp.pdbj.org/pub/emdb/structures/EMD-15073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15073 | HTTPS FTP |
-Validation report
| Summary document |  emd_15073_validation.pdf.gz emd_15073_validation.pdf.gz | 816.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15073_full_validation.pdf.gz emd_15073_full_validation.pdf.gz | 816.4 KB | Display | |
| Data in XML |  emd_15073_validation.xml.gz emd_15073_validation.xml.gz | 22.4 KB | Display | |
| Data in CIF |  emd_15073_validation.cif.gz emd_15073_validation.cif.gz | 29 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15073 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15073 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15073 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15073 | HTTPS FTP |
-Related structure data
| Related structure data |  8a1eMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15073.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15073.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened final cryo-EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
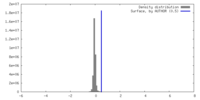
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15073_msk_1.map emd_15073_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
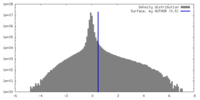
| Density Histograms |
-Mask #2
| File |  emd_15073_msk_2.map emd_15073_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
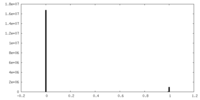
| Density Histograms |
-Half map: Half map A
| File | emd_15073_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
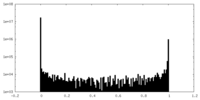
| Density Histograms |
-Half map: Half map B
| File | emd_15073_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rabies virus glycoprotein (chain A) in complex with Fab 1112-1 (c...
| Entire | Name: Rabies virus glycoprotein (chain A) in complex with Fab 1112-1 (chains B and C) and Fab 17C7 (chains D and E) |
|---|---|
| Components |
|
-Supramolecule #1: Rabies virus glycoprotein (chain A) in complex with Fab 1112-1 (c...
| Supramolecule | Name: Rabies virus glycoprotein (chain A) in complex with Fab 1112-1 (chains B and C) and Fab 17C7 (chains D and E) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Rabies lyssavirus Rabies lyssavirus |
| Molecular weight | Theoretical: 330 KDa |
-Macromolecule #1: Glycoprotein
| Macromolecule | Name: Glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rabies virus strain Pasteur vaccin / Strain: Pasteur vaccins / PV Rabies virus strain Pasteur vaccin / Strain: Pasteur vaccins / PV |
| Molecular weight | Theoretical: 56.494551 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KFPIYTIPDK LGPWSPIDIH HLSCPNNLVV EDEGCTNLSG FSYMELKVGY ISAIKMNGFT CTGVVTEAET YTNFVGYVTT TFKRKHFRP TPDACRAAYN WKMAGDPRYE ESLHNPYPDY HWLRTVKTTK ESLVIISPSV ADLDPYDRSL HSPVFPGGNC S GVAVSSTY ...String: KFPIYTIPDK LGPWSPIDIH HLSCPNNLVV EDEGCTNLSG FSYMELKVGY ISAIKMNGFT CTGVVTEAET YTNFVGYVTT TFKRKHFRP TPDACRAAYN WKMAGDPRYE ESLHNPYPDY HWLRTVKTTK ESLVIISPSV ADLDPYDRSL HSPVFPGGNC S GVAVSSTY CSTNHDYTIW MPENPRLGMS CDIFTNSRGK RASKGSETCG FVDERGLYKS LKGACKLKLC GVLGLRLMDG TW VAMQTSN ETKWCPPGQL VNLHDFRSDE IEPLVVEELV KKREECLDAL ESIMTTKSVS FRRLSHLRKL VPGFGKAYTI FNK TLMEAD AHYKSVRTWN EIIPSKGCLR VGGRCHPHVN GVFFNGIILG PDGNVLIPEM QSSLLQQHME LLVSSVIPLM HPLA DPSTV FKNGDEAEDF VEVHLPDVHE RISGVDLGLP NWGKYVLLSA GALTALMLII FLMTCWRRVN RSEPTQHNLR GTGRE VSVT PQSGKIISSW ESHKSGGETR L UniProtKB: Glycoprotein |
-Macromolecule #2: Fab 17C7 heavy chain variable domain
| Macromolecule | Name: Fab 17C7 heavy chain variable domain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.227727 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG VVQPGRSLRL SCAASGFTFS TYAMHWVRQA PGKGLEWVAV VSYDGRTKDY ADSVKGRFTI SRDNSKNTLY LQMNSLRTE DTAVYFCARE RFSGAYFDYW GQGTLVTVSS |
-Macromolecule #3: Fab 17C7 light chain variable domain
| Macromolecule | Name: Fab 17C7 light chain variable domain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.754044 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP GQAPRLLIYD ASNRATGIPA RFSGSGSGTD FTLTISSLEP EDFAVYSCQ QRNNWPPTFG GGTKVEIKRA |
-Macromolecule #4: Fab 1112-1 light chain variable domain
| Macromolecule | Name: Fab 1112-1 light chain variable domain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.668073 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVMTQSHKF MSTSIGDRVS ITCKASQDVS TAVAWYRQKP GQSPKLLIYS ASYRSTGVPD RFTGSGSGTD FTFTISSVQA EDLAVFYCQ QHYSAPLTFG AGTKLELK |
-Macromolecule #5: Fab 1112-1 heavy chain variable domain
| Macromolecule | Name: Fab 1112-1 heavy chain variable domain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.426977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQQPGAE LLKPGTSMKL SCKASGYTFS NYWMHWVKLR PGQGFEWIGE INPFNGGTNF NEKFKSKATL TVDRSSSTAY MQLSSVTSE DSAVYYCTIP LSDYGDWFFA VWGAGTTVTV SS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Blot for 3.5 seconds at -15 N blot force before plunging.. | |||||||||
| Details | Purified sample was concentrated and mixed with 0.07% n-octyl-beta-d-glucoside to final concentrations of 1 mg/mL. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 12884 / Average exposure time: 3.7 sec. / Average electron dose: 44.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Model building was performed with Coot and refined with Phenix. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 108 |
| Output model |  PDB-8a1e: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)