+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fibroblast nucleus sub-volume. | |||||||||
 Map data Map data | sub-volume of EMD-14924. corresponds to Supplementary Movie S6. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleus / chromatin / STEM / DNA | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Sedat J / McDonald A / Elbaum M | |||||||||
| Funding support |  Israel, 1 items Israel, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: A proposed unified interphase nucleus chromosome structure: Preliminary preponderance of evidence. Authors: John Sedat / Angus McDonald / Hu Cang / Joseph Lucas / Muthuvel Arigovindan / Zvi Kam / Cornelis Murre / Michael Elbaum /    Abstract: Cryoelectron tomography of the cell nucleus using scanning transmission electron microscopy and deconvolution processing technology has highlighted a large-scale, 100- to 300-nm interphase chromosome ...Cryoelectron tomography of the cell nucleus using scanning transmission electron microscopy and deconvolution processing technology has highlighted a large-scale, 100- to 300-nm interphase chromosome structure, which is present throughout the nucleus. This study further documents and analyzes these chromosome structures. The paper is divided into four parts: 1) evidence (preliminary) for a unified interphase chromosome structure; 2) a proposed unified interphase chromosome architecture; 3) organization as chromosome territories (e.g., fitting the 46 human chromosomes into a 10-μm-diameter nucleus); and 4) structure unification into a polytene chromosome architecture and lampbrush chromosomes. Finally, the paper concludes with a living light microscopy cell study showing that the G1 nucleus contains very similar structures throughout. The main finding is that this chromosome structure appears to coil the 11-nm nucleosome fiber into a defined hollow structure, analogous to a Slinky helical spring [https://en.wikipedia.org/wiki/Slinky; motif used in Bowerman , 10, e65587 (2021)]. This Slinky architecture can be used to build chromosome territories, extended to the polytene chromosome structure, as well as to the structure of lampbrush chromosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15061.map.gz emd_15061.map.gz | 44.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15061-v30.xml emd-15061-v30.xml emd-15061.xml emd-15061.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15061.png emd_15061.png | 118.9 KB | ||
| Filedesc metadata |  emd-15061.cif.gz emd-15061.cif.gz | 4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15061 http://ftp.pdbj.org/pub/emdb/structures/EMD-15061 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15061 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15061 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15061.map.gz / Format: CCP4 / Size: 63.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15061.map.gz / Format: CCP4 / Size: 63.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sub-volume of EMD-14924. corresponds to Supplementary Movie S6. | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 33.5 Å | ||||||||||||||||||||||||||||||||
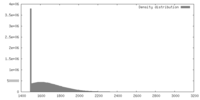
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : WI-38 fibroblast cell
| Entire | Name: WI-38 fibroblast cell |
|---|---|
| Components |
|
-Supramolecule #1: WI-38 fibroblast cell
| Supramolecule | Name: WI-38 fibroblast cell / type: cell / ID: 1 / Parent: 0 / Details: cells cultured directly on gold EM grid |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: lung / Tissue: cell culture Homo sapiens (human) / Organ: lung / Tissue: cell culture |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: Minimal Essential Medium (Gibco) supplemented with 15% fetal calf serum, L-glutamine, and penicillin/streptomycin. |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
| Details | WI-38 embryonic lung fibroblast cells (obtained from Coriell Institute) were cultured directly on gold EM grids and imaged intact by cryo-STEM tomography (CSTET). |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: obtained from L Duchesne (https://doi.org/10.1021/la802876u ) Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Specialist optics | Details: STEM data collection in BF mode. |
| Details | NanoProbe STEM acquisition. |
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number grids imaged: 1 / Number real images: 61 / Average exposure time: 12.6 sec. / Average electron dose: 3.8 e/Å2 / Details: Cryo-STEM tilt series |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 20.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 0.0 µm / Nominal defocus min: 0.0 µm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Gatan STEM BF/ADF detector |
|---|---|
| Final reconstruction | Algorithm: BACK PROJECTION / Software: (Name: TOMO3D, PRIISM/IVE) / Number images used: 56 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)