+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | VWF tubules of D1D2 and D'A1A2A3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
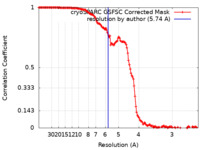
| Method | helical reconstruction / cryo EM / Resolution: 5.74 Å | |||||||||
 Authors Authors | Javitt G / Fass D | |||||||||
| Funding support |  Israel, 1 items Israel, 1 items
| |||||||||
 Citation Citation |  Journal: Blood / Year: 2022 Journal: Blood / Year: 2022Title: Assembly of von Willebrand factor tubules with in vivo helical parameters requires A1 domain insertion. Authors: Gabriel Javitt / Noa Yeshaya / Lev Khmelnitsky / Deborah Fass /  Abstract: The von Willebrand factor (VWF) glycoprotein is stored in tubular form in Weibel-Palade bodies (WPBs) before secretion from endothelial cells into the bloodstream. The organization of VWF in the ...The von Willebrand factor (VWF) glycoprotein is stored in tubular form in Weibel-Palade bodies (WPBs) before secretion from endothelial cells into the bloodstream. The organization of VWF in the tubules promotes formation of covalently linked VWF polymers and enables orderly secretion without polymer tangling. Recent studies have described the high-resolution structure of helical tubular cores formed in vitro by the D1D2 and D'D3 amino-terminal protein segments of VWF. Here we show that formation of tubules with the helical geometry observed for VWF in intracellular WPBs requires also the VWA1 (A1) domain. We reconstituted VWF tubules from segments containing the A1 domain and discovered it to be inserted between helical turns of the tubule, altering helical parameters and explaining the increased robustness of tubule formation when A1 is present. The conclusion from this observation is that the A1 domain has a direct role in VWF assembly, along with its known activity in hemostasis after secretion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15004.map.gz emd_15004.map.gz | 121.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15004-v30.xml emd-15004-v30.xml emd-15004.xml emd-15004.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15004_fsc.xml emd_15004_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15004.png emd_15004.png | 136.4 KB | ||
| Others |  emd_15004_half_map_1.map.gz emd_15004_half_map_1.map.gz emd_15004_half_map_2.map.gz emd_15004_half_map_2.map.gz | 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15004 http://ftp.pdbj.org/pub/emdb/structures/EMD-15004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15004 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15004.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15004.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15004_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15004_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : VWF tubules of D1D2 and D'D3A1 domains
| Entire | Name: VWF tubules of D1D2 and D'D3A1 domains |
|---|---|
| Components |
|
-Supramolecule #1: VWF tubules of D1D2 and D'D3A1 domains
| Supramolecule | Name: VWF tubules of D1D2 and D'D3A1 domains / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 5.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 1.5 sec. / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)