+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Oligomeric structure of SynDLP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BDLP / cyanobacteria / membrane remodeling / LIPID BINDING PROTEIN | |||||||||
| Function / homology | Mitofusin family / Dynamin, N-terminal / Dynamin family / GTPase activity / GTP binding / P-loop containing nucleoside triphosphate hydrolase / membrane / Slr0869 protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
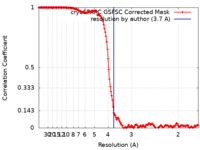
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Gewehr L / Junglas B / Jilly R / Franz J / Wenyu EZ / Weidner T / Bonn M / Sachse C / Schneider D | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: SynDLP is a dynamin-like protein of Synechocystis sp. PCC 6803 with eukaryotic features. Authors: Lucas Gewehr / Benedikt Junglas / Ruven Jilly / Johannes Franz / Wenyu Eva Zhu / Tobias Weidner / Mischa Bonn / Carsten Sachse / Dirk Schneider /   Abstract: Dynamin-like proteins are membrane remodeling GTPases with well-understood functions in eukaryotic cells. However, bacterial dynamin-like proteins are still poorly investigated. SynDLP, the dynamin- ...Dynamin-like proteins are membrane remodeling GTPases with well-understood functions in eukaryotic cells. However, bacterial dynamin-like proteins are still poorly investigated. SynDLP, the dynamin-like protein of the cyanobacterium Synechocystis sp. PCC 6803, forms ordered oligomers in solution. The 3.7 Å resolution cryo-EM structure of SynDLP oligomers reveals the presence of oligomeric stalk interfaces typical for eukaryotic dynamin-like proteins. The bundle signaling element domain shows distinct features, such as an intramolecular disulfide bridge that affects the GTPase activity, or an expanded intermolecular interface with the GTPase domain. In addition to typical GD-GD contacts, such atypical GTPase domain interfaces might be a GTPase activity regulating tool in oligomerized SynDLP. Furthermore, we show that SynDLP interacts with and intercalates into membranes containing negatively charged thylakoid membrane lipids independent of nucleotides. The structural characteristics of SynDLP oligomers suggest it to be the closest known bacterial ancestor of eukaryotic dynamin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14993.map.gz emd_14993.map.gz | 171.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14993-v30.xml emd-14993-v30.xml emd-14993.xml emd-14993.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14993_fsc.xml emd_14993_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14993.png emd_14993.png | 45.7 KB | ||
| Masks |  emd_14993_msk_1.map emd_14993_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14993.cif.gz emd-14993.cif.gz | 6.3 KB | ||
| Others |  emd_14993_half_map_1.map.gz emd_14993_half_map_1.map.gz emd_14993_half_map_2.map.gz emd_14993_half_map_2.map.gz | 317.7 MB 317.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14993 http://ftp.pdbj.org/pub/emdb/structures/EMD-14993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14993 | HTTPS FTP |
-Validation report
| Summary document |  emd_14993_validation.pdf.gz emd_14993_validation.pdf.gz | 743.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14993_full_validation.pdf.gz emd_14993_full_validation.pdf.gz | 742.7 KB | Display | |
| Data in XML |  emd_14993_validation.xml.gz emd_14993_validation.xml.gz | 24.1 KB | Display | |
| Data in CIF |  emd_14993_validation.cif.gz emd_14993_validation.cif.gz | 31.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14993 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14993 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14993 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14993 | HTTPS FTP |
-Related structure data
| Related structure data |  7zw6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14993.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14993.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.869 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14993_msk_1.map emd_14993_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14993_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14993_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : filamentous homo-oligomer of SynDLP
| Entire | Name: filamentous homo-oligomer of SynDLP |
|---|---|
| Components |
|
-Supramolecule #1: filamentous homo-oligomer of SynDLP
| Supramolecule | Name: filamentous homo-oligomer of SynDLP / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Slr0869 protein
| Macromolecule | Name: Slr0869 protein / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 93.705398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKIAPQCQN LREQVNQLIE LLRQEPTLRS QQDTSIVETA LGKALSPRFE IVFAGAFSAG KSMLINALLE RELLYSAEGH ATGTECHIE YANANEERVV LTFLSEAEIR QQALILAKYL NVNVGDLNIN QPEAVKVVSQ YCQKIIAEEG GENKSERAKQ A NALHLLLI ...String: MSKIAPQCQN LREQVNQLIE LLRQEPTLRS QQDTSIVETA LGKALSPRFE IVFAGAFSAG KSMLINALLE RELLYSAEGH ATGTECHIE YANANEERVV LTFLSEAEIR QQALILAKYL NVNVGDLNIN QPEAVKVVSQ YCQKIIAEEG GENKSERAKQ A NALHLLLI GFEQNRERIN TVQNSTYSMD QLNFSSLAEA AGYARRGANS AVLKRLDYFC NHSLLKDGNV LVDLPGIDAP VK EDAERAY RKIESPDTSA VICVLKPAAA GDMSAEETQL LERISKNHGI RDRVFYVFNR IDDTWYNTQL RQRLEGLIQS QFR DNSRVY KTSGLLGFYG SQVKQTNSST RFGLDSIFAT TIKGFDGEEE TPQFVSEFNN YCANSGKLLS TAFRVSVNGY ETSN ENYVR ILSEWGIPLV DQLIHDSGIE SFRSGIGLYL AEEKYPELFA TLANDLQPLC IALRQFYLEN YRQLDSQPRE IAAMK AQEL TLLNQEMQNL GIEFKKYMSA QINDVVIGND REFDQDFTKL KARMVARLDE LLKTFSVMNA YKRATESHPR NSTAPF IAV LVEALYYLAN ELEDAFIEAI HELVKNFFQR LGDRLRKVDC YHQVYRLVGN DGGIEQLLRR AEEDITKALV NEARTEC DR YVRESPRFYD EGTFSIYQFR QTLQQTSQGY DAQAIVEAEP AIKELLKLDF EPKVFNTVRK NFRQTVNNTL KTHLLPMA E EQAQIILEQY DVARKYREQT LEQDAEEKIA RNSRLQSEIK QKIDLYQTSI VSINECLKAM QIFEQLPVIT ESDITKQAE IVADADFVEI VELEHHHHHH UniProtKB: Slr0869 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting force -10 Blotting time 3 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 8322 / Average exposure time: 2.0 sec. / Average electron dose: 26.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 49000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7zw6: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)