[English] 日本語
 Yorodumi
Yorodumi- EMDB-14923: Structure of the human tRNA splicing endonuclease defines substra... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human tRNA splicing endonuclease defines substrate recognition | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNP / endonuclease / tRNA / splicing / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA-intron endonuclease complex / tRNA-type intron splice site recognition and cleavage / tRNA-intron lyase / tRNA-intron lyase activity / tRNA splicing, via endonucleolytic cleavage and ligation / tRNA processing in the nucleus / tRNA processing / mRNA processing / nucleic acid binding / lyase activity ...tRNA-intron endonuclease complex / tRNA-type intron splice site recognition and cleavage / tRNA-intron lyase / tRNA-intron lyase activity / tRNA splicing, via endonucleolytic cleavage and ligation / tRNA processing in the nucleus / tRNA processing / mRNA processing / nucleic acid binding / lyase activity / centrosome / nucleolus / nucleoplasm / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
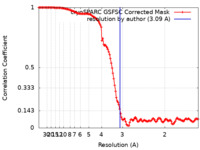
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||
 Authors Authors | Sekulovski S / Trowitzsch S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis of substrate recognition by human tRNA splicing endonuclease TSEN. Authors: Samoil Sekulovski / Lukas Sušac / Lukas S Stelzl / Robert Tampé / Simon Trowitzsch /  Abstract: Heterotetrameric human transfer RNA (tRNA) splicing endonuclease TSEN catalyzes intron excision from precursor tRNAs (pre-tRNAs), utilizing two composite active sites. Mutations in TSEN and its ...Heterotetrameric human transfer RNA (tRNA) splicing endonuclease TSEN catalyzes intron excision from precursor tRNAs (pre-tRNAs), utilizing two composite active sites. Mutations in TSEN and its associated RNA kinase CLP1 are linked to the neurodegenerative disease pontocerebellar hypoplasia (PCH). Despite the essential function of TSEN, the three-dimensional assembly of TSEN-CLP1, the mechanism of substrate recognition, and the structural consequences of disease mutations are not understood in molecular detail. Here, we present single-particle cryogenic electron microscopy reconstructions of human TSEN with intron-containing pre-tRNAs. TSEN recognizes the body of pre-tRNAs and pre-positions the 3' splice site for cleavage by an intricate protein-RNA interaction network. TSEN subunits exhibit large unstructured regions flexibly tethering CLP1. Disease mutations localize far from the substrate-binding interface and destabilize TSEN. Our work delineates molecular principles of pre-tRNA recognition and cleavage by human TSEN and rationalizes mutations associated with PCH. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14923.map.gz emd_14923.map.gz | 203.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14923-v30.xml emd-14923-v30.xml emd-14923.xml emd-14923.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14923_fsc.xml emd_14923_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14923.png emd_14923.png | 43.4 KB | ||
| Masks |  emd_14923_msk_1.map emd_14923_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14923.cif.gz emd-14923.cif.gz | 6.2 KB | ||
| Others |  emd_14923_half_map_1.map.gz emd_14923_half_map_1.map.gz emd_14923_half_map_2.map.gz emd_14923_half_map_2.map.gz | 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14923 http://ftp.pdbj.org/pub/emdb/structures/EMD-14923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14923 | HTTPS FTP |
-Related structure data
| Related structure data |  7zrzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14923.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14923.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.78 Å | ||||||||||||||||||||||||||||||||||||
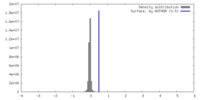
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14923_msk_1.map emd_14923_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
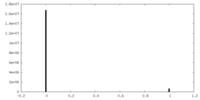
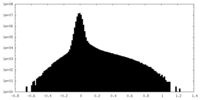
| Density Histograms |
-Half map: half map B
| File | emd_14923_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
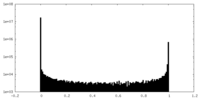
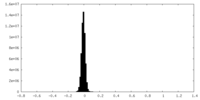
| Density Histograms |
-Half map: half map A
| File | emd_14923_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
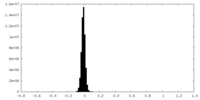
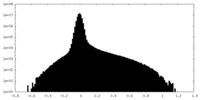
| Density Histograms |
- Sample components
Sample components
-Entire : Human TSEN with pre-tRNA-Arg-TCT
| Entire | Name: Human TSEN with pre-tRNA-Arg-TCT |
|---|---|
| Components |
|
-Supramolecule #1: Human TSEN with pre-tRNA-Arg-TCT
| Supramolecule | Name: Human TSEN with pre-tRNA-Arg-TCT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: tRNA-splicing endonuclease subunit Sen34
| Macromolecule | Name: tRNA-splicing endonuclease subunit Sen34 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: tRNA-intron lyase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.805811 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLVVEVANGR SLVWGAEAVQ ALRERLGVGG RTVGALPRGP RQNSRLGLPL LLMPEEARLL AEIGAVTLVS APRPDSRHHS LALTSFKRQ QEESFQEQSA LAAEARETRR QELLEKITEG QGGSGGSGGS GGSRSALLVQ LATARPRPVK ARPLDWRVQS K DWPHAGRP ...String: MLVVEVANGR SLVWGAEAVQ ALRERLGVGG RTVGALPRGP RQNSRLGLPL LLMPEEARLL AEIGAVTLVS APRPDSRHHS LALTSFKRQ QEESFQEQSA LAAEARETRR QELLEKITEG QGGSGGSGGS GGSRSALLVQ LATARPRPVK ARPLDWRVQS K DWPHAGRP AHELRYSIYR DLWERGFFLS AAGKFGGDFL VYPGDPLRFF AHYIAQCWAP EDTIPLQDLV AAGRLGTSVR KT LLLCSPQ PDGKVVYTSL QWASLQ UniProtKB: tRNA-splicing endonuclease subunit Sen34, tRNA-splicing endonuclease subunit Sen34 |
-Macromolecule #2: tRNA-splicing endonuclease subunit Sen2
| Macromolecule | Name: tRNA-splicing endonuclease subunit Sen2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: tRNA-intron lyase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.165961 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEAVFHAPK RKRRVYETYE SPLPIPFGQD HGPLKEFKIF RAEMINNNVI VRNAEDIEQL YGKGYFGKGI LSRSRPSFTI SGGSGGRIF EYLQLSLEEA FFLVYALGCL SIYYEKEPLT IVKLWKAFTV VQPTFRTTYM AYHYFRSKGW VPKVGLKYGT D LLLYRKGP ...String: MAEAVFHAPK RKRRVYETYE SPLPIPFGQD HGPLKEFKIF RAEMINNNVI VRNAEDIEQL YGKGYFGKGI LSRSRPSFTI SGGSGGRIF EYLQLSLEEA FFLVYALGCL SIYYEKEPLT IVKLWKAFTV VQPTFRTTYM AYHYFRSKGW VPKVGLKYGT D LLLYRKGP PFYFASYSVI IELVDDHFEG SLRRPLSWKS LAALSRVSVN VSKELMLCYL IKPSTMTDKE MESPECMKRI KV QEVILSR WVSSRERSDQ DDL UniProtKB: tRNA-splicing endonuclease subunit Sen2, tRNA-splicing endonuclease subunit Sen2 |
-Macromolecule #3: tRNA-splicing endonuclease subunit Sen54
| Macromolecule | Name: tRNA-splicing endonuclease subunit Sen54 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.551711 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEPEPEPAAV EVPAGRVLSA RELFAARSRS QKLPQRSHGP KDFLPDGSAA QAERLRRCRE ELWQLLAEQR VERLGSLVAA EWRPEEGFV ELKSPAGKFW QTMGFSEQGR QRLHPEEALY LLECGSIHLF HQDLPLSIQE AYQLLLTDHT VTFLQYQVFS H LKRLGYVV ...String: MEPEPEPAAV EVPAGRVLSA RELFAARSRS QKLPQRSHGP KDFLPDGSAA QAERLRRCRE ELWQLLAEQR VERLGSLVAA EWRPEEGFV ELKSPAGKFW QTMGFSEQGR QRLHPEEALY LLECGSIHLF HQDLPLSIQE AYQLLLTDHT VTFLQYQVFS H LKRLGYVV RRFQPSSVLS GGSGGSGGSG GSGSVLQTTH LPDGGARLLE KSGGLEIIFD VYQADAVATF RKNNPGKPYA RM CISGFDE PVPDLCSLKR LSYQSGDVPL IFALVDHGDI SFYSFRDFTL PQDVGH UniProtKB: tRNA-splicing endonuclease subunit Sen54, tRNA-splicing endonuclease subunit Sen54 |
-Macromolecule #4: tRNA-splicing endonuclease subunit Sen15
| Macromolecule | Name: tRNA-splicing endonuclease subunit Sen15 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.860889 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDEKTTGWRG GHVVEGLAGE LEQLRARLEH HPQGQREPGE NLYFQGMEER GDSEPTPGCS GLGPGGVRGF GDGGGAPSWA PEDAWMGTH PKYLEMMELD IGDATQVYVA FLVYLDLMES KSWHEVNCVG LPELQLICLV GTEIEGEGLQ TVVPTPITAS L SHNRIREI ...String: MDEKTTGWRG GHVVEGLAGE LEQLRARLEH HPQGQREPGE NLYFQGMEER GDSEPTPGCS GLGPGGVRGF GDGGGAPSWA PEDAWMGTH PKYLEMMELD IGDATQVYVA FLVYLDLMES KSWHEVNCVG LPELQLICLV GTEIEGEGLQ TVVPTPITAS L SHNRIREI LKASRKLQGD PDLPMSFTLA IVESDSTIVY YKLTDGFMLP DPQNISLRR UniProtKB: tRNA-splicing endonuclease subunit Sen15 |
-Macromolecule #5: pre-tRNA Arg TCT 3-2
| Macromolecule | Name: pre-tRNA Arg TCT 3-2 / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.747045 KDa |
| Sequence | String: GGCUCUGUGG CGCAAUGGAU AGCGCAUUGG ACUUCUAGAU AGUUAGAGAA AUUCAAAGGU UGUGGGUUCG AGUCCCACCA GAGUCGCCA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)