[English] 日本語
 Yorodumi
Yorodumi- EMDB-14883: CryoEM structure of HSP90-CDC37-BRAF(V600E)-PP5(closed) complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of HSP90-CDC37-BRAF(V600E)-PP5(closed) complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of type II interferon-mediated signaling pathway / peptidyl-serine dephosphorylation / positive regulation of nuclear receptor-mediated glucocorticoid signaling pathway / response to arachidonate / HSP90-CDC37 chaperone complex / negative regulation of proteasomal protein catabolic process / Aryl hydrocarbon receptor signalling / aryl hydrocarbon receptor complex / peptidyl-threonine dephosphorylation / CD4-positive, alpha-beta T cell differentiation ...regulation of type II interferon-mediated signaling pathway / peptidyl-serine dephosphorylation / positive regulation of nuclear receptor-mediated glucocorticoid signaling pathway / response to arachidonate / HSP90-CDC37 chaperone complex / negative regulation of proteasomal protein catabolic process / Aryl hydrocarbon receptor signalling / aryl hydrocarbon receptor complex / peptidyl-threonine dephosphorylation / CD4-positive, alpha-beta T cell differentiation / positive regulation of axon regeneration / histone methyltransferase binding / dynein axonemal particle / CD4-positive or CD8-positive, alpha-beta T cell lineage commitment / negative regulation of synaptic vesicle exocytosis / Signalling to p38 via RIT and RIN / receptor ligand inhibitor activity / proximal dendrite / head morphogenesis / positive regulation of type 2 mitophagy / ARMS-mediated activation / endothelial cell apoptotic process / myeloid progenitor cell differentiation / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / protein kinase regulator activity / positive regulation of D-glucose transmembrane transport / regulation of cyclin-dependent protein serine/threonine kinase activity / negative regulation of fibroblast migration / ATP-dependent protein binding / positive regulation of protein localization to cell surface / establishment of protein localization to membrane / mitogen-activated protein kinase kinase kinase binding / regulation of T cell differentiation / positive regulation of axonogenesis / Negative feedback regulation of MAPK pathway / Frs2-mediated activation / post-transcriptional regulation of gene expression / protein dephosphorylation / protein-serine/threonine phosphatase / response to morphine / Respiratory syncytial virus genome replication / stress fiber assembly / telomerase holoenzyme complex assembly / positive regulation of transforming growth factor beta receptor signaling pathway / : / Drug-mediated inhibition of ERBB2 signaling / Resistance of ERBB2 KD mutants to trastuzumab / Resistance of ERBB2 KD mutants to sapitinib / Resistance of ERBB2 KD mutants to tesevatinib / Resistance of ERBB2 KD mutants to neratinib / Resistance of ERBB2 KD mutants to osimertinib / Resistance of ERBB2 KD mutants to afatinib / Resistance of ERBB2 KD mutants to AEE788 / Resistance of ERBB2 KD mutants to lapatinib / Drug resistance in ERBB2 TMD/JMD mutants / Uptake and function of diphtheria toxin / regulation of type I interferon-mediated signaling pathway / TPR domain binding / dendritic growth cone / face development / cellular response to cadmium ion / protein serine/threonine phosphatase activity / MAP kinase kinase activity / Assembly and release of respiratory syncytial virus (RSV) virions / phosphatase activity / thyroid gland development / synaptic vesicle exocytosis / protein phosphatase activator activity / The NLRP3 inflammasome / Sema3A PAK dependent Axon repulsion / somatic stem cell population maintenance / regulation of protein ubiquitination / positive regulation of peptidyl-serine phosphorylation / HSF1-dependent transactivation / protein folding chaperone complex / G-protein alpha-subunit binding / MAP kinase kinase kinase activity / response to unfolded protein / protein serine/threonine kinase inhibitor activity / HSF1 activation / Attenuation phase / negative regulation of endothelial cell apoptotic process / chaperone-mediated protein complex assembly / RHOBTB2 GTPase cycle / axonal growth cone / telomere maintenance via telomerase / postsynaptic modulation of chemical synaptic transmission / protein targeting / Purinergic signaling in leishmaniasis infection / negative regulation of MAPK cascade / positive regulation of stress fiber assembly / sperm end piece / supramolecular fiber organization / heat shock protein binding / ERK1 and ERK2 cascade / DNA polymerase binding / Signaling by ERBB2 / phosphoprotein phosphatase activity / positive regulation of substrate adhesion-dependent cell spreading Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
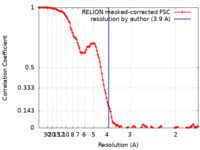
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Oberoi J / Pearl LH | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: HSP90-CDC37-PP5 forms a structural platform for kinase dephosphorylation. Authors: Jasmeen Oberoi / Xavi Aran Guiu / Emily A Outwin / Pascale Schellenberger / Theodoros I Roumeliotis / Jyoti S Choudhary / Laurence H Pearl /  Abstract: Activation of client protein kinases by the HSP90 molecular chaperone system is affected by phosphorylation at multiple sites on HSP90, the kinase-specific co-chaperone CDC37, and the kinase client ...Activation of client protein kinases by the HSP90 molecular chaperone system is affected by phosphorylation at multiple sites on HSP90, the kinase-specific co-chaperone CDC37, and the kinase client itself. Removal of regulatory phosphorylation from client kinases and their release from the HSP90-CDC37 system depends on the Ser/Thr phosphatase PP5, which associates with HSP90 via its N-terminal TPR domain. Here, we present the cryoEM structure of the oncogenic protein kinase client BRAF bound to HSP90-CDC37, showing how the V600E mutation favours BRAF association with HSP90-CDC37. Structures of HSP90-CDC37-BRAF complexes with PP5 in autoinhibited and activated conformations, together with proteomic analysis of its phosphatase activity on BRAF and CRAF, reveal how PP5 is activated by recruitment to HSP90 complexes. PP5 comprehensively dephosphorylates client proteins, removing interaction sites for regulatory partners such as 14-3-3 proteins and thus performing a 'factory reset' of the kinase prior to release. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14883.map.gz emd_14883.map.gz | 119.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14883-v30.xml emd-14883-v30.xml emd-14883.xml emd-14883.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14883_fsc.xml emd_14883_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14883.png emd_14883.png | 82.4 KB | ||
| Filedesc metadata |  emd-14883.cif.gz emd-14883.cif.gz | 7.8 KB | ||
| Others |  emd_14883_half_map_1.map.gz emd_14883_half_map_1.map.gz emd_14883_half_map_2.map.gz emd_14883_half_map_2.map.gz | 106.2 MB 106.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14883 http://ftp.pdbj.org/pub/emdb/structures/EMD-14883 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14883 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14883 | HTTPS FTP |
-Related structure data
| Related structure data |  7zr5MC  7zr0C  7zr6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14883.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14883.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_14883_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14883_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HSP90-CDC37-BRAF(V600E)-PP5 (closed) complex
| Entire | Name: HSP90-CDC37-BRAF(V600E)-PP5 (closed) complex |
|---|---|
| Components |
|
-Supramolecule #1: HSP90-CDC37-BRAF(V600E)-PP5 (closed) complex
| Supramolecule | Name: HSP90-CDC37-BRAF(V600E)-PP5 (closed) complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37 and Seri...
| Supramolecule | Name: Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37 and Serine/threonine-protein kinase B-raf type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Serine/threonine-protein phosphatase 5
| Supramolecule | Name: Serine/threonine-protein phosphatase 5 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Heat shock protein HSP 90-beta
| Macromolecule | Name: Heat shock protein HSP 90-beta / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 86.223469 KDa |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: MRGSHHHHHH HHGMALEVLF QGPSAMPEEV HHGEEEVETF AFQAEIAQLM SLIINTFYSN KEIFLRELIS NASDALDKIR YESLTDPSK LDSGKELKID IIPNPQERTL TLVDTGIGMT KADLINNLGT IAKSGTKAFM EALQAGADIS MIGQFGVGFY S AYLVAEKV ...String: MRGSHHHHHH HHGMALEVLF QGPSAMPEEV HHGEEEVETF AFQAEIAQLM SLIINTFYSN KEIFLRELIS NASDALDKIR YESLTDPSK LDSGKELKID IIPNPQERTL TLVDTGIGMT KADLINNLGT IAKSGTKAFM EALQAGADIS MIGQFGVGFY S AYLVAEKV VVITKHNDDE QYAWESSAGG SFTVRADHGE PIGRGTKVIL HLKEDQTEYL EERRVKEVVK KHSQFIGYPI TL YLEKERE KEISDDEAEE EKGEKEEEDK DDEEKPKIED VGSDEEDDSG KDKKKKTKKI KEKYIDQEEL NKTKPIWTRN PDD ITQEEY GEFYKSLTND WEDHLAVKHF SVEGQLEFRA LLFIPRRAPF DLFENKKKKN NIKLYVRRVF IMDSCDELIP EYLN FIRGV VDSEDLPLNI SREMLQQSKI LKVIRKNIVK KCLELFSELA EDKENYKKFY EAFSKNLKLG IHEDSTNRRR LSELL RYHT SQSGDEMTSL SEYVSRMKET QKSIYYITGE SKEQVANSAF VERVRKRGFE VVYMTEPIDE YCVQQLKEFD GKSLVS VTK EGLELPEDEE EKKKMEESKA KFENLCKLMK EILDKKVEKV TISNRLVSSP CCIVTSTYGW TANMERIMKA QALRDNS TM GYMMAKKHLE INPDHPIVET LRQKAEADKN DKAVKDLVVL LFETALLSSG FSLEDPQTHS NRIYRMIKLG LGIDEDEV A AEEPNAAVPD EIPPLEGDED ASRMEEVD UniProtKB: Heat shock protein HSP 90-beta |
-Macromolecule #2: Hsp90 co-chaperone Cdc37
| Macromolecule | Name: Hsp90 co-chaperone Cdc37 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.853816 KDa |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: MVDYSVWDHI EV(SEP)DDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEG GKA ELERLQAEAQ QLRKEERSWE QKLEEMRKKE KSMPWNVDTL SKDGFSKSMV NTKPEKTEED SEEVREQKHK TFVEKYE KQ IKHFGMLRRW ...String: MVDYSVWDHI EV(SEP)DDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEG GKA ELERLQAEAQ QLRKEERSWE QKLEEMRKKE KSMPWNVDTL SKDGFSKSMV NTKPEKTEED SEEVREQKHK TFVEKYE KQ IKHFGMLRRW DDSQKYLSDN VHLVCEETAN YLVIWCIDLE VEEKCALMEQ VAHQTIVMQF ILELAKSLKV DPRACFRQ F FTKIKTADRQ YMEGFNDELE AFKERVRGRA KLRIEKAMKE YEEEERKKRL GPGGLDPVEV YESLPEELQK CFDVKDVQM LQDAISKMDP TDAKYHMQRC IDSGLWVPNS KASEAKEGEE AGPGDPLLEA VPKTGDEKDV SVLEVLFQGP LEHHHHHHHH UniProtKB: Hsp90 co-chaperone Cdc37 |
-Macromolecule #3: Serine/threonine-protein kinase B-raf
| Macromolecule | Name: Serine/threonine-protein kinase B-raf / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 90.934508 KDa |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: MGGSHHHHHH HHGGSWSHPQ FEKGGGSGGG SGGGSWSHPQ FEKGAETAVP NSLEVLFQGP SAMAALSGGG GGGAEPGQAL FNGDMEPEA GAGAGAAASS AADPAIPEEV WNIKQMIKLT QEHIEALLDK FGGEHNPPSI YLEAYEEYTS KLDALQQREQ Q LLESLGNG ...String: MGGSHHHHHH HHGGSWSHPQ FEKGGGSGGG SGGGSWSHPQ FEKGAETAVP NSLEVLFQGP SAMAALSGGG GGGAEPGQAL FNGDMEPEA GAGAGAAASS AADPAIPEEV WNIKQMIKLT QEHIEALLDK FGGEHNPPSI YLEAYEEYTS KLDALQQREQ Q LLESLGNG TDFSVSSSAS MDTVTSSSSS SLSVLPSSLS VFQNPTDVAR SNPKSPQKPI VRVFLPNKQR TVVPARCGVT VR DSLKKAL MMRGLIPECC AVYRIQDGEK KPIGWDTDIS WLTGEELHVE VLENVPLTTH NFVRKTFFTL AFCDFCRKLL FQG FRCQTC GYKFHQRCST EVPLMCVNYD QLDLLFVSKF FEHHPIPQEE ASLAETALTS GSSPSAPASD SIGPQILTSP SPSK SIPIP QPFRPADEDH RNQFGQRDRS SSAPNVHINT IEPVNIDDLI RDQGFRGDGG STTGLSATPP ASLPGSLTNV KALQK SPGP QRERKSSSSS EDRNRMKTLG RRDSSDDWEI PDGQITVGQR IGSGSFGTVY KGKWHGDVAV KMLNVTAPTP QQLQAF KNE VGVLRKTRHV NILLFMGYST KPQLAIVTQW CEGSSLYHHL HIIETKFEMI KLIDIARQTA QGMDYLHAKS IIHRDLK SN NIFLHEDLTV KIGDFGLATE KSRWSGSHQF EQLSGSILWM APEVIRMQDK NPYSFQSDVY AFGIVLYELM TGQLPYSN I NNRDQIIFMV GRGYLSPDLS KVRSNCPKAM KRLMAECLKK KRDERPLFPQ ILASIELLAR SLPKIHRSAS EPSLNRAGF QTEDFSLYAC ASPKTPIQAG GYGAFPVH UniProtKB: Serine/threonine-protein kinase B-raf |
-Macromolecule #4: Serine/threonine-protein phosphatase 5
| Macromolecule | Name: Serine/threonine-protein phosphatase 5 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: protein-serine/threonine phosphatase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.020387 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DEPPADGALK RAEELKTQAN DYFKAKDYEN AIKFYSQAIE LNPSNAIYYG NRSLAYLRTE CYGYALGDAT RAIELDKKYI KGYYRRAAS NMALGKFRAA LRDYETVVKV KPHDKDAKMK YQECNKIVKQ KAFERAIAGD EHKRSVVDSL DIESMTIEDE Y SGPKLEDG ...String: DEPPADGALK RAEELKTQAN DYFKAKDYEN AIKFYSQAIE LNPSNAIYYG NRSLAYLRTE CYGYALGDAT RAIELDKKYI KGYYRRAAS NMALGKFRAA LRDYETVVKV KPHDKDAKMK YQECNKIVKQ KAFERAIAGD EHKRSVVDSL DIESMTIEDE Y SGPKLEDG KVTISFMKEL MQWYKDQKKL HRKCAYQILV QVKEVLSKLS TLVETTLKET EKITVCGDTH GQFYDLLNIF EL NGLPSET NPYIFNGDFV DRGSFSVEVI LTLFGFKLLY PDHFHLLRGN HETDNMNQIY GFEGEVKAKY TAQMYELFSE VFE WLPLAQ CINGKVLIMH GGLFSEDGVT LDDIRKIERN RQPPDSGPMC DLLWSDPQPQ NGRSISKRGV SCQFGPDVTK AFLE ENNLD YIIRSHEVKA EGYEVAHGGR CVTVFSAPNY CDQMGNKASY IHLQGSDLRP QFHQFTAVPH PNVKPMAYAN TLLQL GMMH HHHHH UniProtKB: Serine/threonine-protein phosphatase 5 |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)