[English] 日本語
 Yorodumi
Yorodumi- EMDB-14802: Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | |||||||||
 Map data Map data | DDB1 deltaBPB/E27/STAT2 CCD main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Interferon / ubiquitin-proteasome system / Cullin-RING ubiquitin ligases (CRL) / DDB1 / DCAFs / viral DCAF (vDCAF) / cytomegalovirus / STAT2 / IRF9 / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationISGF3 complex / positive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / negative regulation of type I interferon-mediated signaling pathway / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / type I interferon-mediated signaling pathway ...ISGF3 complex / positive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / negative regulation of type I interferon-mediated signaling pathway / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / type I interferon-mediated signaling pathway / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / regulation of mitochondrial fission / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / negative regulation of reproductive process / negative regulation of developmental process / ubiquitin-like protein ligase binding / viral release from host cell / cullin family protein binding / cell surface receptor signaling pathway via JAK-STAT / ectopic germ cell programmed cell death / positive regulation of viral genome replication / proteasomal protein catabolic process / positive regulation of gluconeogenesis / nucleotide-excision repair / Recognition of DNA damage by PCNA-containing replication complex / regulation of circadian rhythm / DNA Damage Recognition in GG-NER / Dual Incision in GG-NER / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / Formation of Incision Complex in GG-NER / Wnt signaling pathway / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / positive regulation of protein catabolic process / cellular response to UV / rhythmic process / site of double-strand break / Neddylation / ubiquitin-dependent protein catabolic process / protein-macromolecule adaptor activity / defense response to virus / proteasome-mediated ubiquitin-dependent protein catabolic process / damaged DNA binding / DNA-binding transcription factor activity, RNA polymerase II-specific / chromosome, telomeric region / protein ubiquitination / DNA-binding transcription factor activity / DNA repair / apoptotic process / DNA damage response / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / protein-containing complex binding / nucleolus / positive regulation of transcription by RNA polymerase II / protein-containing complex / extracellular space / DNA binding / extracellular exosome / nucleoplasm / identical protein binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Murid betaherpesvirus 8 / Murid betaherpesvirus 8 /  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.78 Å | |||||||||
 Authors Authors | Lauer S / Spahn CMT / Schwefel D | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor. Authors: Vu Thuy Khanh Le-Trilling / Sofia Banchenko / Darius Paydar / Pia Madeleine Leipe / Lukas Binting / Simon Lauer / Andrea Graziadei / Robin Klingen / Christine Gotthold / Jörg Bürger / ...Authors: Vu Thuy Khanh Le-Trilling / Sofia Banchenko / Darius Paydar / Pia Madeleine Leipe / Lukas Binting / Simon Lauer / Andrea Graziadei / Robin Klingen / Christine Gotthold / Jörg Bürger / Thilo Bracht / Barbara Sitek / Robert Jan Lebbink / Anna Malyshkina / Thorsten Mielke / Juri Rappsilber / Christian Mt Spahn / Sebastian Voigt / Mirko Trilling / David Schwefel /     Abstract: Human cytomegalovirus (CMV) is a ubiquitously distributed pathogen whose rodent counterparts such as mouse and rat CMV serve as common infection models. Here, we conducted global proteome profiling ...Human cytomegalovirus (CMV) is a ubiquitously distributed pathogen whose rodent counterparts such as mouse and rat CMV serve as common infection models. Here, we conducted global proteome profiling of rat CMV-infected cells and uncovered a pronounced loss of the transcription factor STAT2, which is crucial for antiviral interferon signalling. Via deletion mutagenesis, we found that the viral protein E27 is required for CMV-induced STAT2 depletion. Cellular and in vitro analyses showed that E27 exploits host-cell Cullin4-RING ubiquitin ligase (CRL4) complexes to induce poly-ubiquitylation and proteasomal degradation of STAT2. Cryo-electron microscopy revealed how E27 mimics molecular surface properties of cellular CRL4 substrate receptors called DCAFs (DDB1- and Cullin4-associated factors), thereby displacing them from the catalytic core of CRL4. Moreover, structural analyses showed that E27 recruits STAT2 through a bipartite binding interface, which partially overlaps with the IRF9 binding site. Structure-based mutations in M27, the murine CMV homologue of E27, impair the interferon-suppressing capacity and virus replication in mouse models, supporting the conserved importance of DCAF mimicry for CMV immune evasion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14802.map.gz emd_14802.map.gz | 36.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14802-v30.xml emd-14802-v30.xml emd-14802.xml emd-14802.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14802.png emd_14802.png | 172.5 KB | ||
| Filedesc metadata |  emd-14802.cif.gz emd-14802.cif.gz | 8.5 KB | ||
| Others |  emd_14802_half_map_1.map.gz emd_14802_half_map_1.map.gz emd_14802_half_map_2.map.gz emd_14802_half_map_2.map.gz | 35.7 MB 35.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14802 http://ftp.pdbj.org/pub/emdb/structures/EMD-14802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14802 | HTTPS FTP |
-Related structure data
| Related structure data |  7zn7MC  7znnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14802.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14802.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DDB1 deltaBPB/E27/STAT2 CCD main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||
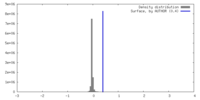
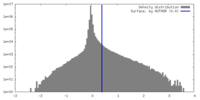
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: DDB1 deltaBPB/E27/STAT2 CCD half map A
| File | emd_14802_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DDB1 deltaBPB/E27/STAT2 CCD half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
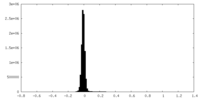
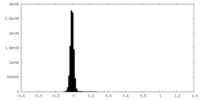
| Density Histograms |
-Half map: DDB1 deltaBPB/E27/STAT2 CCD half map B
| File | emd_14802_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DDB1 deltaBPB/E27/STAT2 CCD half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
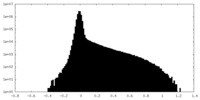
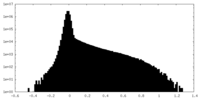
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of RCMV-E E27 with human DDB1 (deltaBPB) and rat STAT2
| Entire | Name: Ternary complex of RCMV-E E27 with human DDB1 (deltaBPB) and rat STAT2 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of RCMV-E E27 with human DDB1 (deltaBPB) and rat STAT2
| Supramolecule | Name: Ternary complex of RCMV-E E27 with human DDB1 (deltaBPB) and rat STAT2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Murid betaherpesvirus 8 Murid betaherpesvirus 8 |
| Molecular weight | Theoretical: 100 KDa |
-Supramolecule #2: DNA damage-binding protein 1
| Supramolecule | Name: DNA damage-binding protein 1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 / Details: delta396-705 GNGNSG |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: E27
| Supramolecule | Name: E27 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Murid betaherpesvirus 8 Murid betaherpesvirus 8 |
-Supramolecule #4: Signal transducer and activator of transcription
| Supramolecule | Name: Signal transducer and activator of transcription / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA damage-binding protein 1
| Macromolecule | Name: DNA damage-binding protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.671461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGMSYNYVV TAQKPTAVNG CVTGHFTSAE DLNLLIAKNT RLEIYVVTAE GLRPVKEVGM YGKIAVMELF RPKGESKDLL FILTAKYNA CILEYKQSGE SIDIITRAHG NVQDRIGRPS ETGIIGIIDP ECRMIGLRLY DGLFKVIPLD RDNKELKAFN I RLEELHVI ...String: GPGMSYNYVV TAQKPTAVNG CVTGHFTSAE DLNLLIAKNT RLEIYVVTAE GLRPVKEVGM YGKIAVMELF RPKGESKDLL FILTAKYNA CILEYKQSGE SIDIITRAHG NVQDRIGRPS ETGIIGIIDP ECRMIGLRLY DGLFKVIPLD RDNKELKAFN I RLEELHVI DVKFLYGCQA PTICFVYQDP QGRHVKTYEV SLREKEFNKG PWKQENVEAE ASMVIAVPEP FGGAIIIGQE SI TYHNGDK YLAIAPPIIK QSTIVCHNRV DPNGSRYLLG DMEGRLFMLL LEKEEQMDGT VTLKDLRVEL LGETSIAECL TYL DNGVVF VGSRLGDSQL VKLNVDSNEQ GSYVVAMETF TNLGPIVDMC VVDLERQGQG QLVTCSGAFK EGSLRIIRNG IGIG NGNSG EIQKLHIRTV PLYESPRKIC YQEVSQCFGV LSSRIEVQDT SGGTTALRPS ASTQALSSSV SSSKLFSSST APHET SFGE EVEVHNLLII DQHTFEVLHA HQFLQNEYAL SLVSCKLGKD PNTYFIVGTA MVYPEEAEPK QGRIVVFQYS DGKLQT VAE KEVKGAVYSM VEFNGKLLAS INSTVRLYEW TTEKELRTEC NHYNNIMALY LKTKGDFILV GDLMRSVLLL AYKPMEG NF EEIARDFNPN WMSAVEILDD DNFLGAENAF NLFVCQKDSA ATTDEERQHL QEVGLFHLGE FVNVFCHGSL VMQNLGET S TPTQGSVLFG TVNGMIGLVT SLSESWYNLL LDMQNRLNKV IKSVGKIEHS FWRSFHTERK TEPATGFIDG DLIESFLDI SRPKMQEVVA NLQYDDGSGM KREATADDLI KVVEELTRIH UniProtKB: DNA damage-binding protein 1, DNA damage-binding protein 1 |
-Macromolecule #2: B27a
| Macromolecule | Name: B27a / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Murid betaherpesvirus 8 / Strain: isolate England Murid betaherpesvirus 8 / Strain: isolate England |
| Molecular weight | Theoretical: 76.341898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGMDLRDRQ CESDRNEDNG ETERQAIKER PNAMKIVFEL IKDETADPFC RKFILDNLVQ LKDIEQAARF GFAIEPGSED YNRGVRSFI RLKEPYNHVE LKMSYLKNAR FATANGAYGI RGLTPAWDSA IWGLLREVEA VPYSNPFTFP TCERLEGILN R LEYRPEAT ...String: GPGMDLRDRQ CESDRNEDNG ETERQAIKER PNAMKIVFEL IKDETADPFC RKFILDNLVQ LKDIEQAARF GFAIEPGSED YNRGVRSFI RLKEPYNHVE LKMSYLKNAR FATANGAYGI RGLTPAWDSA IWGLLREVEA VPYSNPFTFP TCERLEGILN R LEYRPEAT DACRTLCRAT VAVQYAISIL YGYDRSASVP RYLRRLIDEY IDLQDRIRRL GIVPVLKISG KDMNIVQNVL PE RVCAQIG IPFTYEACLL DEYYDCIESN IEQMYDLLCM CKECGMRRME RFERVRYNTI GERYPKKKRD RERFFEENYV IIH SHPDLG VLKLPKIRHL NPLQQLMMCR YLSKDLLYDT QFGPSNAFSR AQGVPLPFSA VQETDMNIAY ALYLASNSYF MCFL LRCVR DVLRHEEEAF RKLILRLVNE AVEIIRDDVN SRVWAGADSP VFVCVDPRES GISAEERDLA IARSLEELTF SNELV EGSH FGGFCRDAAR FLDLIDAFHF PKALREHRAR EDLLLHTFKI EKMYDNRPLS AYYGDRLVPY HVLMGGHRRR DGAVVR TGG VNLTDNEIVR GHWRADDMMN ARLRYFDSID SIEMIRNVSI RQETLMFDLD RMYISRSELE SLESDVESDN ESEVLAG GA CALPDRQSSS EVDSMCD UniProtKB: B27a |
-Macromolecule #3: Signal transducer and activator of transcription
| Macromolecule | Name: Signal transducer and activator of transcription / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.172953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGMAQWEML QNLDSPFLDQ LHELYSQSLL PMDVRQHLAA WIEDQNWREA VLGSDHSKAN MLYFNILDQL NQWDHYSLDS DYLLLQHNL RKFNRDIQMF PNGPTQLAEM IFNLLLEEKR ILNQAQRAQE MQPRPAPEAA GESQQQLEIE TRILDLQTAV E ALVRSIRQ ...String: GPGMAQWEML QNLDSPFLDQ LHELYSQSLL PMDVRQHLAA WIEDQNWREA VLGSDHSKAN MLYFNILDQL NQWDHYSLDS DYLLLQHNL RKFNRDIQMF PNGPTQLAEM IFNLLLEEKR ILNQAQRAQE MQPRPAPEAA GESQQQLEIE TRILDLQTAV E ALVRSIRQ LKDVQDVFSF RYTVLNHKKT SSLDPHQSQQ RQLVQTTANE LDRMRKEVLD ISKALVGRLT TLVDLLLPKL DE WKAQQQK SCIGAPAPEL QLDQLEQWLT AGAKVLFHLR QLLKQLKDMS LMLRYEGDMF GQGVDLQNAQ VTELLQRLLQ RSF VVETQP CMPQTLHRPL ILKTGSKFTV RTRLLVRLQE GNESLKAEVS IDRNSELPGF RKFNILTSNQ KSLTPEEGQR QGLI WDFGF LTLVEQRSVG AGKGSNKGPL PVTEELHIIS AMVEYVYQGL KMKLQTDTLP VVIISNMNQL SIAWASILWF NMLSS TPKD QQFFCKVPKA PWSLLGPALS WQFSSYIGRG LDPEQLGMLR NKLFGKNCKT EDALLSWADF SKRESPPGKI PFWTWL DKI LELVHDHLKD LWNDGRIMGF VSRSEERRLL KKMVSGTFLL RFSETSEGGI TCSWVEHQDD DKVLIYSVQP YTKEVLQ SL PLTEIIRHYQ VLAEENAPEN PLRFLYPRVP RDEAFGCYYQ EKVNLEERRK YLKHRLIVIS NRQADELQQP LELKQDPE S LELDAELMSV MELARDLELQ SMLQVGLDLG VEPKPDPTLS PAPQILLEPD PARALNQLLP EPDLPQDLQQ LNTGEMEIF RNNINIDDVM PYGDPVLAGQ SIIEEAYRSC PSHFDVDGPL IPSED UniProtKB: Signal transducer and activator of transcription |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.13 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: 45 seconds adsorption 2 seconds blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-50 / Number grids imaged: 4 / Number real images: 8069 / Average exposure time: 10.0 sec. / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 31000 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 112 Target criteria: Fast gradient-driven minimization of combined map and restraints target |
| Output model |  PDB-7zn7: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)